Toward a Home Test for COVID-19 Diagnosis: DNA Machine for Amplification-Free SARS-CoV-2 Detection in Clinical Samples
- PMID: 36031581
- PMCID: PMC9538286
- DOI: 10.1002/cmdc.202200382
Toward a Home Test for COVID-19 Diagnosis: DNA Machine for Amplification-Free SARS-CoV-2 Detection in Clinical Samples
Abstract
Nucleic acid-based detection of RNA viruses requires an annealing procedure to obtain RNA/probe or RNA/primer complexes for unwinding stable structures of folded viral RNA. In this study, we designed a protein-enzyme-free nano-construction, named four-armed DNA machine (4DNM), that requires neither an amplification stage nor a high-temperature annealing step for SARS-CoV-2 detection. It uses a binary deoxyribozyme (BiDz) sensor incorporated in a DNA nanostructure equipped with a total of four RNA-binding arms. Additional arms were found to improve the limit of detection at least 10-fold. The sensor distinguished SARS-CoV-2 from other respiratory viruses and correctly identified five positive and six negative clinical samples verified by quantitative polymerase chain reaction (RT-qPCR). The strategy reported here can be used for the detection of long natural RNA and can become a basis for a point-of-care or home diagnostic test.
Keywords: 10-23 DNAzyme; DNA machine; binary hybridization probe; deoxyribozyme sensor; detection of folded RNA.
© 2022 Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
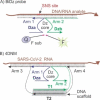
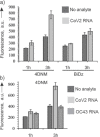

References
-
- Feng W., Newbigging A. M., Le C., Pang B., Peng H., Cao Y., Wu J., Abbas G., Song J., Wang D. B., Cui M., Tao J., Tyrrell D. L., Zhang X. E., Zhang H., Le X. C., Anal. Chem. 2020, 92, 10196–10209. - PubMed
-
- None
-
- Nurputra D. K., Kusumaatmaja A., Hakim M. S., Hidayat S. N., Julian T., Sumanto B., Mahendradhata Y., Saktiawati A. M. I., Wasisto H. S., Triyana K., NPJ Digit. Med. 2022, 5, 115, https://pubmed.ncbi.nlm.nih.gov/35974062/; - PMC - PubMed
-
- Ignaszak A. S., Adv. Mater. Technol. 2022, 2200208, https://pubmed.ncbi.nlm.nih.gov/35942251/; - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

