Prediction of prognosis, immunogenicity and efficacy of immunotherapy based on glutamine metabolism in lung adenocarcinoma
- PMID: 36032135
- PMCID: PMC9403193
- DOI: 10.3389/fimmu.2022.960738
Prediction of prognosis, immunogenicity and efficacy of immunotherapy based on glutamine metabolism in lung adenocarcinoma
Abstract
Background: Glutamine (Gln) metabolism has been reported to play an essential role in cancer. However, a comprehensive analysis of its role in lung adenocarcinoma is still unavailable. This study established a novel system of quantification of Gln metabolism to predict the prognosis and immunotherapy efficacy in lung cancer. Further, the Gln metabolism in tumor microenvironment (TME) was characterized and the Gln metabolism-related genes were identified for targeted therapy.
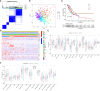
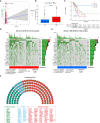
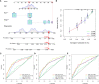
Methods: We comprehensively evaluated the patterns of Gln metabolism in 513 patients diagnosed with lung adenocarcinoma (LUAD) based on 73 Gln metabolism-related genes. Based on differentially expressed genes (DEGs), a risk model was constructed using Cox regression and Lasso regression analysis. The prognostic efficacy of the model was validated using an individual LUAD cohort form Shandong Provincial Hospital, an integrated LUAD cohort from GEO and pan-cancer cohorts from TCGA databases. Five independent immunotherapy cohorts were used to validate the model performance in predicting immunotherapy efficacy. Next, a series of single-cell sequencing analyses were used to characterize Gln metabolism in TME. Finally, single-cell sequencing analysis, transcriptome sequencing, and a series of in vitro experiments were used to explore the role of EPHB2 in LUAD.
Results: Patients with LUAD were eventually divided into low- and high-risk groups. Patients in low-risk group were characterized by low levels of Gln metabolism, survival advantage, "hot" immune phenotype and benefit from immunotherapy. Compared with other cells, tumor cells in TME exhibited the most active Gln metabolism. Among immune cells, tumor-infiltrating T cells exhibited the most active levels of Gln metabolism, especially CD8 T cell exhaustion and Treg suppression. EPHB2, a key gene in the model, was shown to promote LUAD cell proliferation, invasion and migration, and regulated the Gln metabolic pathway. Finally, we found that EPHB2 was highly expressed in macrophages, especially M2 macrophages. It may be involved in the M2 polarization of macrophages and mediate the negative regulation of M2 macrophages in NK cells.
Conclusion: This study revealed that the Gln metabolism-based model played a significant role in predicting prognosis and immunotherapy efficacy in lung cancer. We further characterized the Gln metabolism of TME and investigated the Gln metabolism-related gene EPHB2 to provide a theoretical framework for anti-tumor strategy targeting Gln metabolism.
Keywords: EphB2; glutamine metabolism; immunotherapy; lung adenocarcinoma; prognosis; tumor microenvironment.
Copyright © 2022 Liu, Shen, Gu, Zheng, Wang, Ma and Du.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures











Similar articles
-
Prediction of prognosis and immunotherapy efficacy based on metabolic landscape in lung adenocarcinoma by bulk, single-cell RNA sequencing and Mendelian randomization analyses.Aging (Albany NY). 2024 May 20;16(10):8772-8809. doi: 10.18632/aging.205838. Epub 2024 May 20. Aging (Albany NY). 2024. PMID: 38771130 Free PMC article.
-
Macrophage heterogeneity and oncogenic mechanisms in lung adenocarcinoma: insights from scRNA-seq analysis and predictive modeling.Front Immunol. 2025 Jan 9;15:1491872. doi: 10.3389/fimmu.2024.1491872. eCollection 2024. Front Immunol. 2025. PMID: 39850883 Free PMC article.
-
Development and validation of polyamines metabolism-associated gene signatures to predict prognosis and immunotherapy response in lung adenocarcinoma.Front Immunol. 2023 Jun 2;14:1070953. doi: 10.3389/fimmu.2023.1070953. eCollection 2023. Front Immunol. 2023. PMID: 37334367 Free PMC article.
-
Development of fatty acid metabolism-related models in lung adenocarcinomaA Review.Medicine (Baltimore). 2023 Jan 6;102(1):e32542. doi: 10.1097/MD.0000000000032542. Medicine (Baltimore). 2023. PMID: 36607846 Free PMC article. Review.
-
Identification and characterization of nucleotide metabolism and neuroendocrine regulation-associated modification patterns in stomach adenocarcinoma with auxiliary prognostic assessment and immunotherapy response prediction.Front Endocrinol (Lausanne). 2023 Jan 16;13:1076521. doi: 10.3389/fendo.2022.1076521. eCollection 2022. Front Endocrinol (Lausanne). 2023. PMID: 36726460 Free PMC article. Review.
Cited by
-
Integrated bioinformatics analysis identifies ALDH18A1 as a prognostic hub gene in glutamine metabolism in lung adenocarcinoma.Discov Oncol. 2025 Jan 2;16(1):1. doi: 10.1007/s12672-024-01698-3. Discov Oncol. 2025. PMID: 39747729 Free PMC article.
-
Beyond the tumor: Enhancing pancreatic cancer therapy through glutamine metabolism and innovative drug delivery.J Cell Commun Signal. 2025 Jul 9;19(3):e70033. doi: 10.1002/ccs3.70033. eCollection 2025 Sep. J Cell Commun Signal. 2025. PMID: 40641572 Free PMC article. Review.
-
Telomere-related prognostic signature for survival assessments in lung adenocarcinoma.Transl Cancer Res. 2024 Sep 30;13(9):4520-4533. doi: 10.21037/tcr-24-767. Epub 2024 Sep 27. Transl Cancer Res. 2024. PMID: 39430816 Free PMC article.
-
Plasma metabolomics reveals risk factors for lung adenocarcinoma.Front Oncol. 2024 Mar 19;14:1277206. doi: 10.3389/fonc.2024.1277206. eCollection 2024. Front Oncol. 2024. PMID: 38567154 Free PMC article.
-
Programmed cell death pathways in lung adenocarcinoma: illuminating tumor drug resistance and therapeutic opportunities through single-cell analysis.Discov Oncol. 2024 Dec 23;15(1):828. doi: 10.1007/s12672-024-01736-0. Discov Oncol. 2024. PMID: 39714518 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

