Targeting the Plasmodium falciparum proteome and organelles for potential antimalarial drug candidates
- PMID: 36033316
- PMCID: PMC9398786
- DOI: 10.1016/j.heliyon.2022.e10390
Targeting the Plasmodium falciparum proteome and organelles for potential antimalarial drug candidates
Abstract
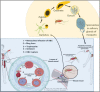
There is an unmet need to unearth alternative treatment options for malaria, wherein this quest is more pressing in recent times due to high morbidity and mortality data arising mostly from the endemic countries coupled with partial diversion of attention from the disease in view of the SARS-Cov-2 pandemic. Available therapeutic options for malaria have been severely threatened with the emergence of resistance to almost all the antimalarial drugs by the Plasmodium falciparum parasite in humans, which is a worrying situation. Artemisinin combination therapies (ACT) that have so far been the mainstay of malaria have encountered resistance by malaria parasite in South East Asia, which is regarded as a notorious ground zero for the emergence of resistance to antimalarial drugs. This review analyzes a few key druggable targets for the parasite and the potential of specific inhibitors to mitigate the emerging antimalarial drug resistance problem by providing a concise assessment of the essential proteins of the malaria parasite that could serve as targets. Moreover, this work provides a summary of the advances made in malaria parasite biology and the potential to leverage these findings for antimalarial drug production.
Keywords: Apical membrane antigen; Dipeptidyl aminopeptidases; Glucose transporters; Malaria; Plasmepsins; Plasmodium rhomboids; Proteases; Schizogony; Subtilisin-like proteins.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Arias M.H., Quiliano M., Bourgeade-Delmas S., Fabing I., Chantal I., Berthier D., Minet C., Eparvier V., Sorres J., Stien D., Galiano S., Aldana I., Valentin A., Garavito G., Deharo E. Alsinol, an arylamino alcohol derivative active against Plasmodium, Babesia, Trypanosoma, and Leishmania: past and new outcomes. Parasitol. Res. 2020;119(10):3503–3515. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

