Potential molecular mechanism in self-renewal is associated with miRNA dysregulation in sacral chordoma - A next-generation RNA sequencing study
- PMID: 36033338
- PMCID: PMC9404356
- DOI: 10.1016/j.heliyon.2022.e10227
Potential molecular mechanism in self-renewal is associated with miRNA dysregulation in sacral chordoma - A next-generation RNA sequencing study
Abstract
Background: Chordoma, the most frequent malignant primary spinal neoplasm, characterized by a high rate of recurrence, is an orphan disease where the clarification of the molecular oncogenesis would be crucial to developing new, effective therapies. Dysregulated expression of non-coding RNAs, especially microRNAs (miRNA) has a significant role in cancer development.
Methods: Next-generation RNA sequencing (NGS) was used for the combinatorial analysis of mRNA-miRNA gene expression profiles in sacral chordoma and nucleus pulposus samples. Advanced bioinformatics workflow was applied to the data to predict miRNA-mRNA regulatory networks with altered activity in chordoma.
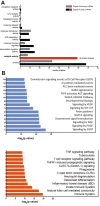
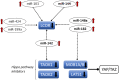
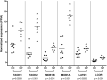
Results: A large set of significantly dysregulated miRNAs in chordoma and their differentially expressed target genes have been identified. Several molecular pathways related to tumorigenesis and the modulation of the immune system are predicted to be dysregulated due to aberrant miRNA expression in chordoma. We identified a gene set including key regulators of the Hippo pathway, which is targeted by differently expressed miRNAs, and validated their altered expression by RT-qPCR. These newly identified miRNA/RNA interactions are predicted to have a role in the self-renewal process of chordoma stem cells, which might sustain the high rate of recurrence for this tumor.
Conclusions: Our results can significantly contribute to the designation of possible targets for the development of anti-chordoma therapies.
Keywords: Chordoma; Next-generation sequencing; Transcriptome profile; mRNA; miRNA.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Szoverfi Z., Lazary A., Bozsodi A., Klemencsics I., Eltes P.E., Varga P.P. Primary spinal tumor mortality score (PSTMS): a novel scoring system for predicting poor survival. Spine J. 2014;14(11):2691–2700. - PubMed
-
- Walcott B.P., Nahed B.V., Mohyeldin A., Coumans J.V., Kahle K.T., Ferreira M.J. Chordoma: current concepts, management, and future directions. Lancet Oncol. 2012;13(2):e69–76. - PubMed
-
- Varga P.P., Szövérfi Z., Fisher C.G., Boriani S., Gokaslan Z.L., Dekutoski M.B., et al. Surgical treatment of sacral chordoma: prognostic variables for local recurrence and overall survival. Eur. Spine J. 2015 May 7;24(5):1092–1101. - PubMed
-
- Yakkioui Y., Temel Y., Creytens D., Jahanshahi A., Fleischeuer R., Santegoeds R.G., et al. A comparison of cell-cycle markers in skull base and sacral chordomas. World Neurosurg. 2014;82(1–2):e311–e318. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

