Molecular and pathological subtypes related to prostate cancer disparities and disease outcomes in African American and European American patients
- PMID: 36033462
- PMCID: PMC9399459
- DOI: 10.3389/fonc.2022.928357
Molecular and pathological subtypes related to prostate cancer disparities and disease outcomes in African American and European American patients
Abstract
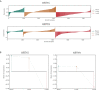
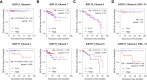
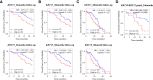
Prostate cancer (PCa) disproportionately affects African American (AA) men, yet present biomarkers do not address the observed racial disparity. The objective of this study was to identify biomarkers with potential benefits to AA PCa patients. Differentially expressed genes (DEG) analysis coupled with gene set enrichment analysis (GSEA) and leading-edge genes analysis showed that the keratin family of genes, including KRT8, KRT15, KRT19, KRT34, and KRT80, constituted the single most prominent family of genes enriched in AA compared to European American (EA) PCa cell lines. In PCa patients (TCGA and MSKCC patient cohorts), KRT8, KRT15, and KRT19 expression were relatively higher in AA than in EA patients. The differences in the expression of KRT15 and KRT19, but not KRT8, were enhanced by Gleason score and ERG fusion status; in low Gleason (Gleason ≤ 6 [TCGA cohort] and Gleason ≤ 7 [MSKCC cohort]), the expression of KRT15 and KRT19 was significantly (p ≤ 0.05) higher in AA than in EA patients. Survival analysis revealed that high expression of KRT15 and KRT19 was associated with increased risk of biochemical recurrence in low Gleason category patients in the TCGA patient cohort. Interestingly, KRT15 and KRT19 expression were also associated with an increased risk of death in the metastatic prostate adenocarcinoma cohort, suggesting the potential to predict the risks of disease recurrence and death in the low Gleason category and advanced disease conditions respectively. Gene set enrichment analysis revealed known oncogenic gene signatures, including KRAS and ERBB2, to be enriched in patients expressing high KRT15 and KRT19. Furthermore, high KRT15 and KRT19 were linked to the basal and LumA PCa subtypes, which are associated with poor postoperative androgen deprivation therapy (ADT) response compared to the LumB subtype. Taken together, the present study identifies genes with high expression in AA than in EA PCa. The identified genes are linked to oncogenic gene signatures, including KRAS and ERBB2, and to basal and LumA PCa subtypes that are associated with poor postoperative ADT response. This study, therefore, reveals biomarkers with the potential to address biomarker bias in PCa risk stratification and/or prognosis.
Keywords: cancer disparities; molecular subtype classification; oncogenic pathways; prognosis; prostate cancer.
Copyright © 2022 Mori, White, Elhussin, Duduyemi, Karanam, Yates and Wang.
Conflict of interest statement
CY is a shareholder in Riptide biosciences and is a consultant in QED Therapeutics, Riptide Biosciences, and Amgen. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Giri VN, Ruth K, Hughes L, Uzzo RG, Chen DYT, Boorjian SA, et al. Racial differences in prediction of time to prostate cancer diagnosis in a prospective screening cohort of high-risk men: effect of TMPRSS2 Met160Val. BJU Int (2011) 107(3):466–70. doi: 10.1111/j.1464-410X.2010.09522.x - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

