B.1.1.7 (Alpha) variant is the most antigenic compared to Wuhan strain, B.1.351, B.1.1.28/triple mutant and B.1.429 variants
- PMID: 36033846
- PMCID: PMC9411949
- DOI: 10.3389/fmicb.2022.895695
B.1.1.7 (Alpha) variant is the most antigenic compared to Wuhan strain, B.1.351, B.1.1.28/triple mutant and B.1.429 variants
Abstract
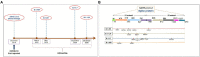
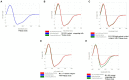
The rapid spread of the SARS-CoV-2 virus and its variants has created a catastrophic impact worldwide. Several variants have emerged, including B.1.351 (Beta), B.1.1.28/triple mutant (P.1), B.1.1.7 (Alpha), and B.1.429 (Epsilon). We performed comparative and comprehensive antigenicity mapping of the total S-glycoprotein using the Wuhan strain and the other variants and identified 9-mer, 15-mer, and 20-mer CTL epitopes through in silico analysis. The study found that 9-mer CTL epitope regions in the B.1.1.7 variant had the highest antigenicity and an average of the three epitope types. Cluster analysis of the 9-mer CTL epitopes depicted one significant cluster at the 70% level with two nodes (KGFNCYFPL and EGFNCYFPL). The phage-displayed peptides showed mimic 9-mer CTL epitopes with three clusters. CD spectra analysis showed the same band pattern of S-glycoprotein of Wuhan strain and all variants other than B.1.429. The developed 3D model of the superantigen (SAg)-like regions found an interaction pattern with the human TCR, indicating that the SAg-like component might interact with the TCR beta chain. The present study identified another partial SAg-like region (ANQFNSAIGKI) from the S-glycoprotein. Future research should examine the molecular mechanism of antigen processing for CD8+ T cells, especially all the variants' antigens of S-glycoprotein.
Keywords: B.1.1.7; CD spectra; CTL epitopes; SAg-like region; antigenicity; cluster; emerging variants; phage-displayed peptides.
Copyright © 2022 Bhattacharya, Sharma, Mallick, Lee, Seo and Chakraborty.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
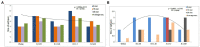








References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

