Where do T cell subsets stand in SARS-CoV-2 infection: an update
- PMID: 36034704
- PMCID: PMC9399648
- DOI: 10.3389/fcimb.2022.964265
Where do T cell subsets stand in SARS-CoV-2 infection: an update
Abstract
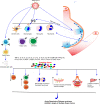
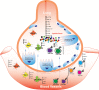
An outbreak of coronavirus disease 2019 (COVID-19) emerged in China in December 2019 and spread so rapidly all around the globe. It's continued and spreading more dangerously in India and Brazil with higher mortality rate. Understanding of the pathophysiology of COVID-19 depends on unraveling of interactional mechanism of SARS-CoV-2 and human immune response. The immune response is a complex process, which can be better understood by understanding the immunological response and pathological mechanisms of COVID-19, which will provide new treatments, increase treatment efficacy, and decrease mortality associated with the disease. In this review we present a amalgamate viewpoint based on the current available knowledge on COVID-19 which includes entry of the virus and multiplication of virus, its pathological effects on the cellular level, immunological reaction, systemic and organ presentation. T cells play a crucial role in controlling and clearing viral infections. Several studies have now shown that the severity of the COVID-19 disease is inversely correlated with the magnitude of the T cell response. Understanding SARS-CoV-2 T cell responses is of high interest because T cells are attractive vaccine targets and could help reduce COVID-19 severity. Even though there is a significant amount of literature regarding SARS-CoV-2, there are still very few studies focused on understanding the T cell response to this novel virus. Nevertheless, a majority of these studies focused on peripheral blood CD4+ and CD8+ T cells that were specific for viruses. The focus of this review is on different subtypes of T cell responses in COVID-19 patients, Th17, follicular helper T (TFH), regulatory T (Treg) cells, and less classical, invariant T cell populations, such as δγ T cells and mucosal-associated invariant T (MAIT) cells etc that could influence disease outcome.
Keywords: COVID-19; T cell; immune response; immunological reaction; pathophysiology.
Copyright © 2022 Tarique, Suhail, Naz, Muhammad, Tabrez, Zughaibi, Abuzenadah, Hashem, Shankar, Saini and Sharma.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Balachandran H., Phetsouphanh C., Agapiou D., Adhikari A., Rodrigo C., Hammoud M., et al. . (2022). Maintenance of broad neutralizing antibodies and memory b cells 1 year post-infection is predicted by SARS-CoV-2-specific CD4+ T cell responses. Cell Rep. 38, 110345. doi: 10.1016/j.celrep.2022.110345 - DOI - PMC - PubMed
-
- Barathan M., Mohamed R., Vadivelu J., Chang L. Y., Saeidi A., Yong Y. K., et al. . (2016). Peripheral loss of CD8(+) CD161(++) TCRValpha7.2(+) mucosal-associated invariant T cells in chronic hepatitis c virus-infected patients. Eur. J. Clin. Invest. 46, 170–180. doi: 10.1111/eci.12581 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

