An inflammation-related signature could predict the prognosis of patients with kidney renal clear cell carcinoma
- PMID: 36035192
- PMCID: PMC9405188
- DOI: 10.3389/fgene.2022.866696
An inflammation-related signature could predict the prognosis of patients with kidney renal clear cell carcinoma
Abstract
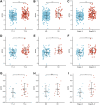
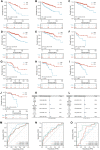
Background: Kidney renal clear cell carcinoma (KIRC) is an inflammation-related carcinoma, and inflammation has been recognized as an important factor in inducing carcinogenesis. To further explore the role of inflammation in KIRC, we developed an inflammation-related signature and verified its correlation with the tumor micro-environment. Methods: After the differential inflammation-related prognostic genes were screened by Lasso regression, the inflammation-related signature (IRS) was constructed based on the risk score of multivariate Cox regression. Then, the prognostic value of the IRS was evaluated by Kaplan-Meier analysis, receiver operating characteristic (ROC) curve analysis and multivariate Cox regression. Gene set variation analysis (GSVA) was applied to screen out enriched signaling pathways. Infiltrated immune cells, tumor mutational burden (TMB) and immune checkpoints were explored by CIBERSORTx and maftool. Results: Four genes (TIMP1, PLAUR, CCL22, and IL15RA) were used to construct the IRS in patients with KIRC. Kaplan-Meier analysis and multivariate Cox regression identified that the IRS could independently predict the prognosis of patients with KIRC in the training and validation groups. The diagnostic value of the nomogram increased from 0.811 to 0.845 after adding the IRS to the multiparameter ROC analysis. The GSVA results indicated that IRS was closely related to primary immunodeficiency and antigen processing and presentation. The immune checkpoint LAG3 was highly expressed in patients with high-risk score (p < 0.05), while CD274 (PD-L1) and HAVCR2 were highly expressed in patients with low-risk score (p < 0.001). There was a significant positive correlation between the high-risk score group and CD8+ T, activated CD4+ memory T, gamma and delta regulatory T and M0 macrophage cells, while the low-risk score group was negatively associated with B memory, plasma, resting CD4+ memory T, activated NK, M1 macrophages and resting mast cells. Conclusion: We found that the IRS might serve as a biomarker to predict the survival of KIRC. Moreover, patients with high or low-risk score might be sensitive to immune drugs at different immune checkpoints.
Keywords: immune checkpoint; immune infiltration; inflammation; renal clear cell carcinoma; signature.
Copyright © 2022 Yu, Zhang, Feng, Li, Xia and Gan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Borrelli C., Ricci B., Vulpis E., Fionda C., Ricciardi M. R., Petrucci M. T., et al. (2018). Drug-induced senescent multiple myeloma cells elicit NK cell proliferation by direct or exosome-mediated IL15 trans-presentation. Cancer Immunol. Res. 6 (7), 860–869. 10.1158/2326-6066.Cir-17-0604 - DOI - PubMed
-
- De Mattia E., Polesel J., Roncato R., Labriet A., Bignucolo A., Gagno S., et al. (2021). IL15RA and SMAD3 genetic variants predict overall survival in metastatic colorectal cancer patients treated with folfiri therapy: A new paradigm. Cancers (Basel) 13 (7), 1705. 10.3390/cancers13071705 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

