Mechanisms of Sporicidal Activity Induced by Ionized Hydrogen Peroxide in the Spores of Bacillus atrophaeus
- PMID: 36035543
- PMCID: PMC9134323
- DOI: 10.1089/apb.20.0060
Mechanisms of Sporicidal Activity Induced by Ionized Hydrogen Peroxide in the Spores of Bacillus atrophaeus
Abstract
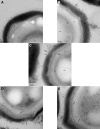
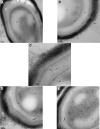
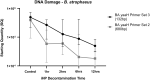
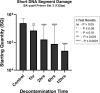
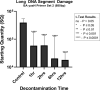
Introduction: Ionized hydrogen peroxide (iHP) is a new technology used for the decontamination of surfaces or laboratory areas. It utilizes a low concentration of hydrogen peroxide (H2O2) mixed with air and ionized through a cold plasma arc. This technology generates reactive oxygen species as a means of decontamination. Objectives: The purpose of this study is to review the effects of iHP on the structure of the spores of Bacillus atrophaeus by observing its effects using transmission electron microscopy (TEM) and also by evaluating the existence of DNA damage by fluorescence-based quantitative polymerase chain reaction (qPCR). Methods: Spore samples of B. atrophaeus decontaminated using iHP at different exposure times (Control, 1, 2, 6, and 12 h) were fixed for TEM. In addition, DNA was extracted for evaluation of DNA damages using fluorescence-based qPCR assays. Results: Damages to the spore structures of B. atrophaeus caused by the decontamination process with iHP at different exposure times (Control, 1, 2, 6, and 12 h) can be observed in micrographs. The effects of the decontamination to short DNA segment (132 base pairs [bp]) of the yaaH gene using qPCR present a linear degradation, and for the long DNA segment (680 bp), it presents a biphasic mode. Conclusion: The results of the qPCR analysis show two initial stages of damage to DNA with very noticeable damage at 12 h contact time, which confirms the observations of the TEM micrographs for the B. atrophaeus spores. The study demonstrates damage to the spore core DNA.
Keywords: Bacillus atrophaeus; DNA damage; decontamination; ionized hydrogen peroxide; transmission electron microscopy.
Copyright 2021, ABSA International 2021.
Conflict of interest statement
No competing financial interests exist.
Figures

References
-
- Gaunt LF, Beggs CB, Georghiou GE. Bactericidal action of the reactive species produced by gas-discharge nonthermal plasma at atmospheric pressure: a review. IEEE Trans Plasma Sci. 2006;34(4):1257–1269.
-
- Das K, Roychoudhury A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front Environ Sci. 2014;2:53.
-
- Kim JY, Lee IH, Kim D, et al. Effects of reactive oxygen species on the biological, structural, and optical properties of Cordyceps pruinosa spores. RSC Advances. 2016;6(36):30699–30709.
-
- Berryman MA, Rodewald RD. An enhanced method for post-embedding immunocytochemical staining which preserves cell membranes. J Histochem Cytochem. 1990;38(2):159–170. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
