Akkermansia muciniphila upregulates genes involved in maintaining the intestinal barrier function via ADP-heptose-dependent activation of the ALPK1/TIFA pathway
- PMID: 36036242
- PMCID: PMC9427033
- DOI: 10.1080/19490976.2022.2110639
Akkermansia muciniphila upregulates genes involved in maintaining the intestinal barrier function via ADP-heptose-dependent activation of the ALPK1/TIFA pathway
Abstract
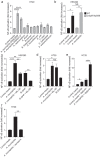
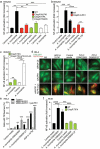
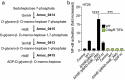
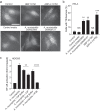
The commensal bacteria that make up the gut microbiota impact the health of their host on multiple levels. In particular, the interactions taking place between the microbe-associated molecule patterns (MAMPs) and pattern recognition receptors (PRRs), expressed by intestinal epithelial cells (IECs), are crucial for maintaining intestinal homeostasis. While numerous studies showed that TLRs and NLRs are involved in the control of gut homeostasis by commensal bacteria, the role of additional innate immune receptors remains unclear. Here, we seek for novel MAMP-PRR interactions involved in the beneficial effect of the commensal bacterium Akkermansia muciniphila on intestinal homeostasis. We show that A. muciniphila strongly activates NF-κB in IECs by releasing one or more potent activating metabolites into the microenvironment. By using drugs, chemical and gene-editing tools, we found that the released metabolite(s) enter(s) epithelial cells and activate(s) NF-κB via an ALPK1, TIFA and TRAF6-dependent pathway. Furthermore, we show that the released molecule has the biological characteristics of the ALPK1 ligand ADP-heptose. Finally, we show that A. muciniphila induces the expression of the MUC2, BIRC3 and TNFAIP3 genes involved in the maintenance of the intestinal barrier function and that this process is dependent on TIFA. Altogether, our data strongly suggest that the commensal A. muciniphila promotes intestinal homeostasis by activating the ALPK1/TIFA/TRAF6 axis, an innate immune pathway exclusively described so far in the context of Gram-negative bacterial infections.
Keywords: ALPK1; Microbiota; NF-κB; akkermansia muciniphila; intestinal epithelial cells.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
-
- Naito T, Mulet C, De Castro C, Molinaro A, Saffarian A, Nigro G, Bérard M, Clerc M, Pedersen AB, Sansonetti PJ, Pédron T, et al. Lipopolysaccharide from crypt-specific core microbiota modulates the colonic epithelial proliferation-to-differentiation balance. mBio. 2017; 8(5):e01680–17. - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
