Molecular analysis reveals a distinct subgenogroup of porcine epidemic diarrhea virus in northern Vietnam in 2018-2019
- PMID: 36036306
- PMCID: PMC9421642
- DOI: 10.1007/s00705-022-05580-x
Molecular analysis reveals a distinct subgenogroup of porcine epidemic diarrhea virus in northern Vietnam in 2018-2019
Abstract
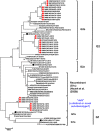
The spike protein (S) of porcine epidemic diarrhea virus (PEDV), in particular, the C-terminal domain of the S1 subunit (S1-CTD), which contains the conserved CO-26K-equivalent (COE) region (aa 499-638), which is recognized by neutralizing antibodies, exhibits a high degree of genetic and antigenic diversity. We analyzed 61 PEDV S1-CTD sequences (630 nt), including 26 from samples collected from seven provinces in northern Vietnam from 2018 to 2019 and 35 other sequences, representing the G1a and 1b, G2a and 2b, and recombinant (G1c) genotypes and vaccines. The majority (73.1%) of the strains (19/26) belonged to subgroup G2b. In a phylogenetic analysis, seven strains were clustered into an independent, distinct subgenogroup named dsG with strong nodal support (98%), separate from both G1a and G1b as well as G2a, 2b, and G1c. Sequence analysis revealed distinct changes (513T>S, 520G>D, 527V>(L/M), 591L>F, 669A>(S/P), and 691V>I) in the COE and S1D regions that were only identified in these Vietnamese strains. This cluster is a new antigenic variant subgroup, and further studies are required to investigate the antigenicity of these variants. The results of this study demonstrated the continuous evolution in the S1 region of Vietnamese PEDV strains, which emphasizes the need for frequent updates of vaccines for effective protection.
© 2022. The Author(s), under exclusive licence to Springer-Verlag GmbH Austria, part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Puranaveja S, Poolperm P, Lertwatcharasarakul P, Kesdaengsakonwut S, Boonsoongnern A, Urairong K, Kitikoon P, Choojai P, Kedkovid R, Teankum K, Thanawongnuwech R. Chinese-like strain of porcine epidemic diarrhea virus, Thailand. Emerg Infect Dis. 2009;15(7):1112–1115. doi: 10.3201/eid1507.081256. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

