Chemical and enzymatic modifications of 5-methylcytosine at the intersection of DNA damage, repair, and epigenetic reprogramming
- PMID: 36037209
- PMCID: PMC9423628
- DOI: 10.1371/journal.pone.0273509
Chemical and enzymatic modifications of 5-methylcytosine at the intersection of DNA damage, repair, and epigenetic reprogramming
Abstract
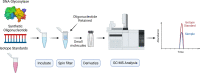
The DNA of all living organisms is persistently damaged by endogenous reactions including deamination and oxidation. Such damage, if not repaired correctly, can result in mutations that drive tumor development. In addition to chemical damage, recent studies have established that DNA bases can be enzymatically modified, generating many of the same modified bases. Irrespective of the mechanism of formation, modified bases can alter DNA-protein interactions and therefore modulate epigenetic control of gene transcription. The simultaneous presence of both chemically and enzymatically modified bases in DNA suggests a potential intersection, or collision, between DNA repair and epigenetic reprogramming. In this paper, we have prepared defined sequence oligonucleotides containing the complete set of oxidized and deaminated bases that could arise from 5-methylcytosine. We have probed these substrates with human glycosylases implicated in DNA repair and epigenetic reprogramming. New observations reported here include: SMUG1 excises 5-carboxyuracil (5caU) when paired with A or G. Both TDG and MBD4 cleave 5-formyluracil and 5caU when mispaired with G. Further, TDG not only removes 5-formylcytosine and 5-carboxycytosine when paired with G, but also when mispaired with A. Surprisingly, 5caU is one of the best substrates for human TDG, SMUG1 and MBD4, and a much better substrate than T. The data presented here introduces some unexpected findings that pose new questions on the interactions between endogenous DNA damage, repair, and epigenetic reprogramming pathways.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

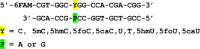



References
-
- Lindahl T, Nyberg B. Heat-induced deamination of cytosine residues in DNA. Biochemistry. 1974;13: 3405–3410. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

