Diploid-dominant life cycles characterize the early evolution of Fungi
- PMID: 36037379
- PMCID: PMC9457484
- DOI: 10.1073/pnas.2116841119
Diploid-dominant life cycles characterize the early evolution of Fungi
Abstract
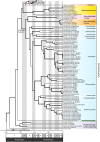
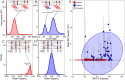
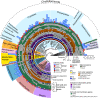
Most of the described species in kingdom Fungi are contained in two phyla, the Ascomycota and the Basidiomycota (subkingdom Dikarya). As a result, our understanding of the biology of the kingdom is heavily influenced by traits observed in Dikarya, such as aerial spore dispersal and life cycles dominated by mitosis of haploid nuclei. We now appreciate that Fungi comprises numerous phylum-level lineages in addition to those of Dikarya, but the phylogeny and genetic characteristics of most of these lineages are poorly understood due to limited genome sampling. Here, we addressed major evolutionary trends in the non-Dikarya fungi by phylogenomic analysis of 69 newly generated draft genome sequences of the zoosporic (flagellated) lineages of true fungi. Our phylogeny indicated five lineages of zoosporic fungi and placed Blastocladiomycota, which has an alternation of haploid and diploid generations, as branching closer to the Dikarya than to the Chytridiomyceta. Our estimates of heterozygosity based on genome sequence data indicate that the zoosporic lineages plus the Zoopagomycota are frequently characterized by diploid-dominant life cycles. We mapped additional traits, such as ancestral cell-cycle regulators, cell-membrane- and cell-wall-associated genes, and the use of the amino acid selenocysteine on the phylogeny and found that these ancestral traits that are shared with Metazoa have been subject to extensive parallel loss across zoosporic lineages. Together, our results indicate a gradual transition in the genetics and cell biology of fungi from their ancestor and caution against assuming that traits measured in Dikarya are typical of other fungal lineages.
Keywords: aquatic fungi; life cycle evolution; phylogenomics; plesiomorphy.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- James T. Y., Berbee M. L., No jacket required--new fungal lineage defies dress code: Recently described zoosporic fungi lack a cell wall during trophic phase. BioEssays 34, 94–102 (2012). - PubMed
-
- Anderson J. B., Kohn L. M., “Dikaryons, diploids, and evolution” in Sex in Fungi: Molecular Determination and Evolutionary Implications, Heitman J., Kronstad J. W., Taylor J., Casselton L., Eds. (ASM Press, 2007), pp. 333–348.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

