Constructing custom-made radiotranscriptomic signatures of vascular inflammation from routine CT angiograms: a prospective outcomes validation study in COVID-19
- PMID: 36038496
- PMCID: PMC9417284
- DOI: 10.1016/S2589-7500(22)00132-7
Constructing custom-made radiotranscriptomic signatures of vascular inflammation from routine CT angiograms: a prospective outcomes validation study in COVID-19
Abstract
Background: Direct evaluation of vascular inflammation in patients with COVID-19 would facilitate more efficient trials of new treatments and identify patients at risk of long-term complications who might respond to treatment. We aimed to develop a novel artificial intelligence (AI)-assisted image analysis platform that quantifies cytokine-driven vascular inflammation from routine CT angiograms, and sought to validate its prognostic value in COVID-19.
Methods: For this prospective outcomes validation study, we developed a radiotranscriptomic platform that uses RNA sequencing data from human internal mammary artery biopsies to develop novel radiomic signatures of vascular inflammation from CT angiography images. We then used this platform to train a radiotranscriptomic signature (C19-RS), derived from the perivascular space around the aorta and the internal mammary artery, to best describe cytokine-driven vascular inflammation. The prognostic value of C19-RS was validated externally in 435 patients (331 from study arm 3 and 104 from study arm 4) admitted to hospital with or without COVID-19, undergoing clinically indicated pulmonary CT angiography, in three UK National Health Service (NHS) trusts (Oxford, Leicester, and Bath). We evaluated the diagnostic and prognostic value of C19-RS for death in hospital due to COVID-19, did sensitivity analyses based on dexamethasone treatment, and investigated the correlation of C19-RS with systemic transcriptomic changes.
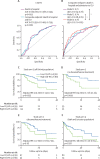
Findings: Patients with COVID-19 had higher C19-RS than those without (adjusted odds ratio [OR] 2·97 [95% CI 1·43-6·27], p=0·0038), and those infected with the B.1.1.7 (alpha) SARS-CoV-2 variant had higher C19-RS values than those infected with the wild-type SARS-CoV-2 variant (adjusted OR 1·89 [95% CI 1·17-3·20] per SD, p=0·012). C19-RS had prognostic value for in-hospital mortality in COVID-19 in two testing cohorts (high [≥6·99] vs low [<6·99] C19-RS; hazard ratio [HR] 3·31 [95% CI 1·49-7·33], p=0·0033; and 2·58 [1·10-6·05], p=0·028), adjusted for clinical factors, biochemical biomarkers of inflammation and myocardial injury, and technical parameters. The adjusted HR for in-hospital mortality was 8·24 (95% CI 2·16-31·36, p=0·0019) in patients who received no dexamethasone treatment, but 2·27 (0·69-7·55, p=0·18) in those who received dexamethasone after the scan, suggesting that vascular inflammation might have been a therapeutic target of dexamethasone in COVID-19. Finally, C19-RS was strongly associated (r=0·61, p=0·00031) with a whole blood transcriptional module representing dysregulation of coagulation and platelet aggregation pathways.
Interpretation: Radiotranscriptomic analysis of CT angiography scans introduces a potentially powerful new platform for the development of non-invasive imaging biomarkers. Application of this platform in routine CT pulmonary angiography scans done in patients with COVID-19 produced the radiotranscriptomic signature C19-RS, a marker of cytokine-driven inflammation driving systemic activation of coagulation and responsible for adverse clinical outcomes, which predicts in-hospital mortality and might allow targeted therapy.
Funding: Engineering and Physical Sciences Research Council, British Heart Foundation, Oxford BHF Centre of Research Excellence, Innovate UK, NIHR Oxford Biomedical Research Centre, Wellcome Trust, Onassis Foundation.
Copyright © 2022 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of Interests CA, KC, CS, and SN are founders, shareholders, and directors of Caristo Diagnostics, a CT image analysis company. CS is a full-time employee and MS is a part-time employee of Caristo diagnostics. JD is shareholder and chair of the advisory board of Caristo Diagnostics. EKO is a consultant and minor shareholder of Caristo Diagnostics. The technology described in this work is subject to patent US10,695,023B2 and patent applications PCT/GB2017/053262, GB2018/1818049.7, GR20180100490, and GR20180100510, licensed through exclusive license to Caristo Diagnostics. Caristo Diagnostics and the authors linked to it have no further conflicts of interest, beyond the above. JD is CMO of Our Future Health; Senior Advisor for Cardiovascular Disease Prevention, NHS Healthcheck Expert Scientific and Clinical Advisory Panel; and Chair of the Review of the National Health Check Programme for Public Health England. JCLR received a Research for Patient Benefit Grant from NIHR, and consulting fees from HeartFlow for physician services. DAd received support from Leicester NIHR Biomedical Research Unit and Innovate UK; grants and contracts from the Medical Research Council; and has two patents issued (Cardiac assist device: EP3277337A1; and angioplasty of calcified arteries: PCT/GB2017/050877) outside the scope of the current study. All other authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- TG/16/3/32687/BHF_/British Heart Foundation/United Kingdom
- 28051/CRUK_/Cancer Research UK/United Kingdom
- MC_UU_00008/5/MRC_/Medical Research Council/United Kingdom
- 203141/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- TG/19/2/34831/BHF_/British Heart Foundation/United Kingdom
- CH/F/21/90009/BHF_/British Heart Foundation/United Kingdom
- MC_PC_19025/MRC_/Medical Research Council/United Kingdom
- 29034/CRUK_/Cancer Research UK/United Kingdom
- MC_PC_19059/MRC_/Medical Research Council/United Kingdom
- CH/09/002/26360/BHF_/British Heart Foundation/United Kingdom
- FS/16/15/32047/BHF_/British Heart Foundation/United Kingdom
- 26988/CRUK_/Cancer Research UK/United Kingdom
- CH/16/1/32013/BHF_/British Heart Foundation/United Kingdom
- RG/F/21/110040/BHF_/British Heart Foundation/United Kingdom
- COV/GLA/20/05/CSO_/Chief Scientist Office/United Kingdom
- 27723/CRUK_/Cancer Research UK/United Kingdom
- MC_UU_00008/6/MRC_/Medical Research Council/United Kingdom
- RE/18/3/34214/BHF_/British Heart Foundation/United Kingdom
- MR/V010182/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

