HnRNPK maintains single strand RNA through controlling double-strand RNA in mammalian cells
- PMID: 36038571
- PMCID: PMC9424213
- DOI: 10.1038/s41467-022-32537-0
HnRNPK maintains single strand RNA through controlling double-strand RNA in mammalian cells
Abstract
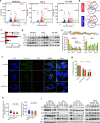
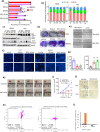
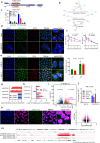
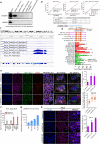
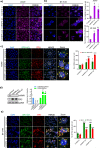
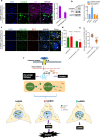
Although antisense transcription is a widespread event in the mammalian genome, double-stranded RNA (dsRNA) formation between sense and antisense transcripts is very rare and mechanisms that control dsRNA remain unknown. By characterizing the FGF-2 regulated transcriptome in normal and cancer cells, we identified sense and antisense transcripts IER3 and IER3-AS1 that play a critical role in FGF-2 controlled oncogenic pathways. We show that IER3 and IER3-AS1 regulate each other's transcription through HnRNPK-mediated post-transcriptional regulation. HnRNPK controls the mRNA stability and colocalization of IER3 and IER3-AS1. HnRNPK interaction with IER3 and IER3-AS1 determines their oncogenic functions by maintaining them in a single-stranded form. hnRNPK depletion neutralizes their oncogenic functions through promoting dsRNA formation and cytoplasmic accumulation. Intriguingly, hnRNPK loss-of-function and gain-of-function experiments reveal its role in maintaining global single- and double-stranded RNA. Thus, our data unveil the critical role of HnRNPK in maintaining single-stranded RNAs and their physiological functions by blocking RNA-RNA interactions.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

