Tumor immune contexture is a determinant of anti-CD19 CAR T cell efficacy in large B cell lymphoma
- PMID: 36038629
- PMCID: PMC9499856
- DOI: 10.1038/s41591-022-01916-x
Tumor immune contexture is a determinant of anti-CD19 CAR T cell efficacy in large B cell lymphoma
Abstract
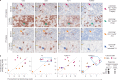
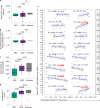
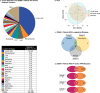
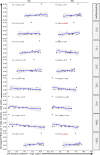
Axicabtagene ciloleucel (axi-cel) is an anti-CD19 chimeric antigen receptor (CAR) T cell therapy approved for relapsed/refractory large B cell lymphoma (LBCL) and has treatment with similar efficacy across conventional LBCL subtypes. Toward patient stratification, we assessed whether tumor immune contexture influenced clinical outcomes after axi-cel. We evaluated the tumor microenvironment (TME) of 135 pre-treatment and post-treatment tumor biopsies taken from 51 patients in the ZUMA-1 phase 2 trial. We uncovered dynamic patterns that occurred within 2 weeks after axi-cel. The biological associations among Immunoscore (quantification of tumor-infiltrating T cell density), Immunosign 21 (expression of pre-defined immune gene panel) and cell subsets were validated in three independent LBCL datasets. In the ZUMA-1 trial samples, clinical response and overall survival were associated with pre-treatment immune contexture as characterized by Immunoscore and Immunosign 21. Circulating CAR T cell levels were associated with post-treatment TME T cell exhaustion. TME enriched for chemokines (CCL5 and CCL22), γ-chain receptor cytokines (IL-15, IL-7 and IL-21) and interferon-regulated molecules were associated with T cell infiltration and markers of activity. Finally, high density of regulatory T cells in pre-treatment TME associated with reduced axi-cel-related neurologic toxicity. These findings advance the understanding of LBCL TME characteristics associated with clinical responses to anti-CD19 CAR T cell therapy and could foster biomarker development and treatment optimization for patients with LBCL.
© 2022. The Author(s).
Conflict of interest statement
N.S. reports employment with Kite, a Gilead company, and stock or other ownership in Gilead Sciences, Bristol Myers Squibb and Seattle Genetics. R.P. reports employment with HalioDx. F.L.L. reports a scientific advisory role for Kite, a Gilead company, Novartis, Amgen, Celgene/Bristol Myers Squibb, GammaDelta Therapeutics, Wugen, Calibr and Allogene; a consultancy with grant options for Cellular Biomedicine Group; and research support from Kite, a Gilead company. M.D.J. reports a consultancy/advisory for Kite, a Gilead company, Novartis, Bristol Myers Squibb and Takeda. S.T. reports employment with, a leadership role at, stock or other ownership in and travel support from HalioDx. C.D. reports stock or other ownership in HalioDx. E.C.C., J.J.K., A.X., M.M. and J.M.R. report employment with Kite, a Gilead company, and stock or other ownership in Gilead Sciences. S.S.N. has received personal fees from Kite, a Gilead company, Merck, Bristol Myers Squibb, Novartis, Celgene, Pfizer, Allogene Therapeutics, Cell Medica/Kuur, Incyte, Precision Biosciences, Legend Biotech, Adicet Bio, Calibr and Unum Therapeutics; has received research support from Kite, a Gilead company, Bristol Myers Squibb, Merck, Poseida, Cellectis, Celgene, Karus Therapeutics, Unum Therapeutics, Allogene Therapeutics, Precision Biosciences and Acerta; has received royalties from Takeda; and has intellectual property related to cell therapy. D.B.M. reports a consultancy or advisory role for Kite-Gilead, Novartis, Juno-Celgene-Bristol Myers Squibb, Adaptive Biotech, Pharmacyclics and Janssen; research funding from Kite-Gilead, Novartis, Juno-Celgene-Bristol Myers Squibb, Adaptive Biotech and Pharmacyclics; patents, royalties or other intellectual property from Pharmacyclics; and travel support from Kite-Gilead, Novartis, Juno-Celgene-Bristol Myers Squibb, Adaptive Biotech, Pharmacyclics and Janssen. C.A.J. reports honoraria from Kite, a Gilead company, Celgene, Novartis, Pfizer, Humanigen, Precision Biosciences, Nkarta, Lonza and AbbVie; a consulting or advisory role with Kite, a Gilead company, Celgene, Novartis, Pfizer, Humanigen, Precision Biosciences, Nkarta, Lonza and AbbVie; speakers bureau participation for Axis and Clinical Care Options; research funding from Pfizer; and travel, accommodations and expenses from Kite, a Gilead company, Celgene, Novartis, Pfizer, Humanigen, Precision Biosciences and Lonza. L.J.L. has no relevant financial relationships to disclose. Y.L. reports a consultancy or advisory role for Kite, a Gilead company, Janssen, Novartis, Celgene, Bluebird Bio, Juno, Legend, Sorrento, Gamida Cells and Vineti; and research funding from Kite, a Gilead company, Janssen, Celgene, Bluebird Bio, Merck and Takeda. A.G. reports a consultancy or advisory role for Kite, a Gilead company, Amgen, Atara, Wugen and Celgene; and honoraria from Kite, a Gilead company. J.C. reports employment with Gilead Sciences; stock or other ownership in Five Prime Therapeutics and Gilead Sciences; patents, royalties or other intellectual property from Five Prime Therapeutics; and travel support from Kite, a Gilead company. V.P. reports employment with, and travel support from, Kite, a Gilead company; and stock or other ownership in Gilead Sciences. Z.W. reports previous employment with Seattle Genetics and current employment with Kite, a Gilead company; and stock or other ownership with Seattle Genetics and Gilead Sciences. A.B. reports employment with Kite, a Gilead company; and stock or other ownership in, a consultancy or advisory role for and travel support from Gilead Sciences. J.G. reports stock or other ownership in HalioDx; honoraria from, and a consultancy or advisory role for, HalioDx, Bristol Myers Squibb, Merck Serono, IO Biotech, Illumina, Northwest Biotherapeutics, Amgen, Catalym, Lunaphore and Merck; research funding from MedImmune, AstraZeneca, IO Biotech, Janssen, Imcheck Therapeutics and PerkinElmer; and patents, royalties or other intellectual property from INSERM. Immunoscore is a registered trademark from INSERM, licensed to HalioDx.
Figures











Comment in
-
In CAR T cell-treated lymphomas, the T cell rich get richer.Nat Med. 2022 Sep;28(9):1757-1758. doi: 10.1038/s41591-022-01922-z. Nat Med. 2022. PMID: 36038630 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

