Human ureteric bud organoids recapitulate branching morphogenesis and differentiate into functional collecting duct cell types
- PMID: 36038632
- PMCID: PMC9957856
- DOI: 10.1038/s41587-022-01429-5
Human ureteric bud organoids recapitulate branching morphogenesis and differentiate into functional collecting duct cell types
Abstract
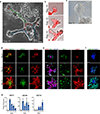
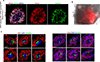
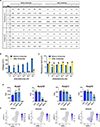
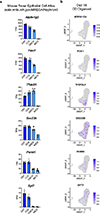
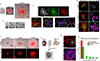
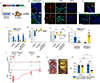
Directed differentiation of human pluripotent stem cells (hPSCs) into functional ureteric and collecting duct (CD) epithelia is essential to kidney regenerative medicine. Here we describe highly efficient, serum-free differentiation of hPSCs into ureteric bud (UB) organoids and functional CD cells. The hPSCs are first induced into pronephric progenitor cells at 90% efficiency and then aggregated into spheres with a molecular signature similar to the nephric duct. In a three-dimensional matrix, the spheres form UB organoids that exhibit branching morphogenesis similar to the fetal UB and correct distal tip localization of RET expression. Organoid-derived cells incorporate into the UB tips of the progenitor niche in chimeric fetal kidney explant culture. At later stages, the UB organoids differentiate into CD organoids, which contain >95% CD cell types as estimated by single-cell RNA sequencing. The CD epithelia demonstrate renal electrophysiologic functions, with ENaC-mediated vectorial sodium transport by principal cells and V-type ATPase proton pump activity by FOXI1-induced intercalated cells.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing financial interests
Dr. Bonventre is an inventor on kidney organoid patents assigned to Mass General Brigham (MGB), and is cofounder and holds equity in Goldfinch Bio. Dr. McCracken has kidney organoid patents pending. Dr. Bonventre’s interests were reviewed and are managed by BWH and MGB in accordance with their conflict-of-interest policies. The other authors have no competing financial interests to declare.
Figures








Comment in
-
Building a kidney tree: Functional collecting duct from human pluripotent stem cells.Dev Cell. 2022 Oct 10;57(19):2251-2253. doi: 10.1016/j.devcel.2022.09.009. Dev Cell. 2022. PMID: 36220079
-
Functional renal collecting ducts from human PSCs.Cell Stem Cell. 2022 Nov 3;29(11):1510-1512. doi: 10.1016/j.stem.2022.10.006. Cell Stem Cell. 2022. PMID: 36332567
References
-
- Takasato M et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

