Proteome-wide analysis of lysine 2-hydroxyisobutyrylation in Frankliniella occidentalis
- PMID: 36038823
- PMCID: PMC9422105
- DOI: 10.1186/s12864-022-08841-w
Proteome-wide analysis of lysine 2-hydroxyisobutyrylation in Frankliniella occidentalis
Abstract
Background: Lysine 2-hydroxyisobutyrylation (Khib) is a novel and conserved post-translational modification (PTM). Frankliniella occidentalis are economically important agricultural pests globally and also notorious for vectoring destructive plant viruses. To better study the disease transmission mechanism of F. occidentalis, it is necessary to conduct in-depth analysis of it. So far, no Khib modification of insects has been reported.
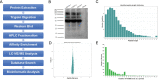
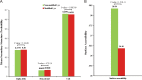
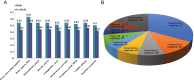
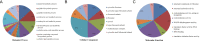
Results: In this study, a proteome-wide analysis of Khib modifications in F. occidentalis was analyzed for the first time through the combination of high performance liquid chromatography fractionation technology and 2-hydroxyisobutyrylated peptide enrichment and other advanced technologies, 4093 Khib sites were identified on 1125 modified proteins. Bioinformatics and functional enrichment analyses showed that Khib-modified proteins were significantly enriched in many cell compartments and pathways, especially related to various cellular components and biological processes, and were more concentrated in ribosomes and proteasome subunits, involved in energy metabolism, protein synthesis and degradation, compared to the other nine species including Japonica rice, Homo sapiens, P. patens, Botrytis, Ustilaginoidea virens, Saccharomyces cerevisiae, T. gondii, C. albicans, and F. oxysporum. And Khib sites on virus-interacting insect proteins were discovered for the first time, such as cyclophilin and endoCP-GN.
Conclusions: After three repeated experiments, we found a total of 4093 Khib sites on 1125 proteins. These modified proteins are mainly concentrated in ribosomes and proteasome subunits, and are widely involved in a variety of critical biological activities and metabolic processes of F. occidentalis. In addition, for the first time, Khib modification sites are found on the proteome of F. occidentalis, and these sites could be acted as for the virus interaction, including cyclophilin and endoCP-GN. The global map of 2-hydroxyisobutyrylation in thrips is an invaluable resource to better understand the biological processes of thrips and provide new means for disease control and mitigation of pest damage to crops.
Keywords: Frankliniella occidentalis; Lysine 2-hydroxyisobutyrylation; Pathogenicity; Post-translational modifications; Proteome.
© 2022. The Author(s).
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures


Similar articles
-
Proteome-Wide Analysis of Lysine 2-Hydroxyisobutyrylation in Aspergillus niger in Peanuts.Front Microbiol. 2021 Aug 18;12:719337. doi: 10.3389/fmicb.2021.719337. eCollection 2021. Front Microbiol. 2021. PMID: 34489910 Free PMC article.
-
Proteome-Wide Analysis of Lysine 2-Hydroxyisobutyrylation in Candida albicans.mSystems. 2021 Feb 2;6(1):e01129-20. doi: 10.1128/mSystems.01129-20. mSystems. 2021. PMID: 33531408 Free PMC article.
-
Comprehensive analysis of lysine lactylation in Frankliniella occidentalis.Front Genet. 2022 Nov 1;13:1014225. doi: 10.3389/fgene.2022.1014225. eCollection 2022. Front Genet. 2022. PMID: 36386791 Free PMC article.
-
Metabolic Functions of Lysine 2-Hydroxyisobutyrylation.Cureus. 2020 Aug 11;12(8):e9651. doi: 10.7759/cureus.9651. Cureus. 2020. PMID: 32923251 Free PMC article. Review.
-
A global invasion by the thrip, Frankliniella occidentalis: Current virus vector status and its management.Insect Sci. 2020 Aug;27(4):626-645. doi: 10.1111/1744-7917.12721. Epub 2019 Oct 23. Insect Sci. 2020. PMID: 31453663 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
- ZR202103070049/Shandong Provincial Natural Science Foundation Project
- ZR202103070049/Shandong Provincial Natural Science Foundation Project
- ZR202103070049/Shandong Provincial Natural Science Foundation Project
- 110202001033 (LS-02) and 110202101045 (LS-05)/China National Tobacco Corporation Green Tobacco Prevention and Control Major Special Project
- 110202001033 (LS-02) and 110202101045 (LS-05)/China National Tobacco Corporation Green Tobacco Prevention and Control Major Special Project
- 110202001033 (LS-02) and 110202101045 (LS-05)/China National Tobacco Corporation Green Tobacco Prevention and Control Major Special Project
- 110202001033 (LS-02) and 110202101045 (LS-05)/China National Tobacco Corporation Green Tobacco Prevention and Control Major Special Project
- SCYC202008/Sichuan Tobacco Company Science and Technology Project
- SCYC202008/Sichuan Tobacco Company Science and Technology Project
- 2020410300270076/Henan Tobacco Company Luoyang City Company Science and Technology Project
- 2020410300270076/Henan Tobacco Company Luoyang City Company Science and Technology Project
- 2020530000241011/Yunnan Tobacco Company Science and Technology Project
- 2020530000241011/Yunnan Tobacco Company Science and Technology Project
LinkOut - more resources
Full Text Sources

