Insights into Systems for Iron-Sulfur Cluster Biosynthesis in Acidophilic Microorganisms
- PMID: 36039043
- PMCID: PMC9628965
- DOI: 10.4014/jmb.2206.06045
Insights into Systems for Iron-Sulfur Cluster Biosynthesis in Acidophilic Microorganisms
Abstract
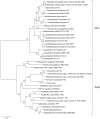
Fe-S clusters are versatile and essential cofactors that participate in multiple and fundamental biological processes. In Escherichia coli, the biogenesis of these cofactors requires either the housekeeping Isc pathway, or the stress-induced Suf pathway which plays a general role under conditions of oxidative stress or iron limitation. In the present work, the Fe-S cluster assembly Isc and Suf systems of acidophilic Bacteria and Archaea, which thrive in highly oxidative environments, were studied. This analysis revealed that acidophilic microorganisms have a complete set of genes encoding for a single system (either Suf or Isc). In acidophilic Proteobacteria and Nitrospirae, a complete set of isc genes (iscRSUAX-hscBA-fdx), but not genes coding for the Suf system, was detected. The activity of the Isc system was studied in Leptospirillum sp. CF-1 (Nitrospirae). RT-PCR experiments showed that eight candidate genes were co-transcribed and conform the isc operon in this strain. Additionally, RT-qPCR assays showed that the expression of the iscS gene was significantly up-regulated in cells exposed to oxidative stress imposed by 260 mM Fe2(SO4)3 for 1 h or iron starvation for 3 h. The activity of cysteine desulfurase (IscS) in CF-1 cell extracts was also up-regulated under such conditions. Thus, the Isc system from Leptospirillum sp. CF-1 seems to play an active role in stressful environments. These results contribute to a better understanding of the distribution and role of Fe-S cluster protein biogenesis systems in organisms that thrive in extreme environmental conditions.
Keywords: Fe-S cluster; Isc; Leptospirillum; Suf; acidophiles; iron starvation; oxidative stress.
Conflict of interest statement
The authors have no financial conflicts of interest to declare.
Figures


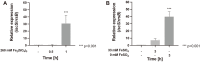
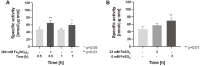
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

