DJ-1 is an essential downstream mediator in PINK1/parkin-dependent mitophagy
- PMID: 36039535
- PMCID: PMC9762950
- DOI: 10.1093/brain/awac313
DJ-1 is an essential downstream mediator in PINK1/parkin-dependent mitophagy
Abstract
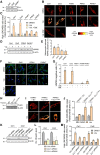
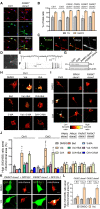
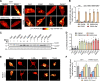
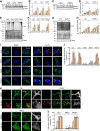
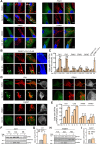
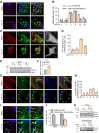
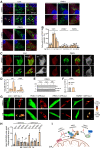
Loss-of-function mutations in the PRKN, PINK1 and PARK7 genes (encoding parkin, PINK1 and DJ-1, respectively) cause autosomal recessive forms of Parkinson's disease. PINK1 and parkin jointly mediate selective autophagy of damaged mitochondria (mitophagy), but the mechanisms by which loss of DJ-1 induces Parkinson's disease are not well understood. Here, we investigated PINK1/parkin-mediated mitophagy in cultured human fibroblasts and induced pluripotent stem cell-derived neurons with homozygous PARK7 mutations. We found that DJ-1 is essential for PINK1/parkin-mediated mitophagy. Loss of DJ-1 did not interfere with PINK1 or parkin activation after mitochondrial depolarization but blocked mitophagy further downstream by inhibiting recruitment of the selective autophagy receptor optineurin to depolarized mitochondria. By contrast, starvation-induced, non-selective autophagy was not affected by loss of DJ-1. In wild-type fibroblasts and induced pluripotent stem cell-derived dopaminergic neurons, endogenous DJ-1 translocated to depolarized mitochondria in close proximity to optineurin. DJ-1 translocation to depolarized mitochondria was dependent on PINK1 and parkin and did not require oxidation of cysteine residue 106 of DJ-1. Overexpression of DJ-1 did not rescue the mitophagy defect of PINK1- or parkin-deficient cells. These findings position DJ-1 downstream of PINK1 and parkin in the same pathway and suggest that disruption of PINK1/parkin/DJ-1-mediated mitophagy is a common pathogenic mechanism in autosomal recessive Parkinson's disease.
Keywords: DJ-1; Parkinson’s disease; autophagy; mitophagy; optineurin.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Guarantors of Brain.
Figures

Comment in
-
Strengthening the link between mitophagy and Parkinson's disease.Brain. 2022 Dec 19;145(12):4154-4156. doi: 10.1093/brain/awac405. Brain. 2022. PMID: 36319596 Free PMC article.
References
-
- Poewe W, Seppi K, Tanner CM, et al. . Parkinson disease. Nat Rev Dis Primers. 2017;3:17013. - PubMed
-
- Malpartida AB, Williamson M, Narendra DP, Wade-Martins R, Ryan BJ. Mitochondrial dysfunction and mitophagy in Parkinson’s disease: From mechanism to therapy. Trends Biochem Sci. 2021;46:329–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

