Spike mutations contributing to the altered entry preference of SARS-CoV-2 omicron BA.1 and BA.2
- PMID: 36039901
- PMCID: PMC9542985
- DOI: 10.1080/22221751.2022.2117098
Spike mutations contributing to the altered entry preference of SARS-CoV-2 omicron BA.1 and BA.2
Abstract
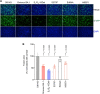
SARS-CoV-2 B.1.1.529.1 (Omicron BA.1) emerged in November 2021 and quickly became the predominant circulating SARS-CoV-2 variant globally. Omicron BA.1 contains more than 30 mutations in the spike protein, which contribute to its altered virological features when compared to the ancestral SARS-CoV-2 or previous SARS-CoV-2 variants. Recent studies by us and others demonstrated that Omicron BA.1 is less dependent on transmembrane serine protease 2 (TMPRSS2), less efficient in spike cleavage, less fusogenic, and adopts an altered propensity to utilize the plasma membrane and endosomal pathways for virus entry. Ongoing studies suggest that these virological features of Omicron BA.1 are in part retained by the subsequent Omicron sublineages. However, the exact spike determinants that contribute to these altered features of Omicron remain incompletely understood. In this study, we investigated the spike determinants for the observed virological characteristics of Omicron. By screening for the individual changes on Omicron BA.1 and BA.2 spike, we identify that 69-70 deletion, E484A, and H655Y contribute to the reduced TMPRSS2 usage while 25-27 deletion, S375F, and T376A result in less efficient spike cleavage. Among the shared spike mutations of BA.1 and BA.2, S375F and H655Y reduce spike-mediated fusogenicity. Interestingly, the H655Y change consistently reduces serine protease usage while increases the use of endosomal proteases. In keeping with these findings, the H655Y substitution alone reduces plasma membrane entry and facilitates endosomal entry when compared to SARS-CoV-2 WT. Overall, our study identifies key changes in Omicron spike that contributes to our understanding on the virological determinant and pathogenicity of Omicron.
Keywords: Omicron BA.1 and BA.2; SARS-CoV-2; endosomal entry pathway; entry; fusogenicity; pathogenesis; spike protein cleavage.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
