In silico analysis and a comparative genomics approach to predict pathogenic trehalase genes in the complete genome of Antarctica Shigella sp. PAMC28760
- PMID: 36040103
- PMCID: PMC9450901
- DOI: 10.1080/21505594.2022.2117679
In silico analysis and a comparative genomics approach to predict pathogenic trehalase genes in the complete genome of Antarctica Shigella sp. PAMC28760
Abstract
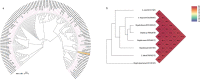
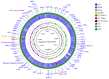
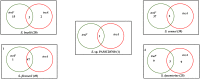
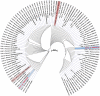
Although four Shigella species (S. flexneri, S. sonnei, S. dysenteriae, and S. boydii) have been reported, S. sp. PAMC 28760, an Antarctica isolate, is the only one with a complete genome deposited in NCBI database as an uncharacterized isolate. Because it is the world's driest, windiest, and coldest continent, Antarctica provides an unfavourable environment for microorganisms. Computational analysis of genomic sequences of four Shigella species and our uncategorized Antarctica isolates Shigella sp. PAMC28760 was performed using MP3 (offline version) program to predict trehalase encoding genes as a pathogenic or non-pathogenic form. Additionally, we employed RAST and Prokka (offline version) annotation programs to determine locations of periplasmic (treA) and cytoplasmic (treF) trehalase genes in studied genomes. Our results showed that only 56 out of 134 Shigella strains had two different trehalase genes (treF and treA). It was revealed that the treF gene tends to be prevalent in Shigella species. In addition, both treA and treF genes were present in our strain S. sp. PAMC28760. The main objective of this study was to predict the prevalence of two different trehalase genes (treF and treA) in the complete genome of Shigella sp. PAMC28760 and other complete genomes of Shigella species. Till date, it is the first study to show that two types of trehalase genes are involved in Shigella species, which could offer insight on how the bacteria use accessible carbohydrate like glucose produced from the trehalose degradation pathway, and importance of periplasmic trehalase involvement in bacterial virulence.
Keywords: HMM; MP3; SVM; Shigella sp.; prokka; trehalase.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
-
- Kotloff KL, Nataro JP, Blackwelder WC, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382(9888):209–222. DOI: 10.1016/S0140-6736(13)60844-2. - DOI - PubMed
-
- Muthuirulandi Sethuvel DP, Devanga Ragupathi NK, Anandan S, et al. Update on: shigella new serogroups/serotypes and their antimicrobial resistance. Lett Appl Microbiol. 2017;64(1):8–18. - PubMed
-
- Lampel KA, Maurelli AT. Shigella species. Ch 11. In: Miliotis M and Bier J, editors. International handbook of foodborne pathogens. Boca Raton: CRC Press. 2003:167–180. doi: 10.1201/9780203912065. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
