SARS-CoV-2 variant spike and accessory gene mutations alter pathogenesis
- PMID: 36040867
- PMCID: PMC9477415
- DOI: 10.1073/pnas.2204717119
SARS-CoV-2 variant spike and accessory gene mutations alter pathogenesis
Abstract
The ongoing COVID-19 pandemic is a major public health crisis. Despite the development and deployment of vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the pandemic persists. The continued spread of the virus is largely driven by the emergence of viral variants, which can evade the current vaccines through mutations in the spike protein. Although these differences in spike are important in terms of transmission and vaccine responses, these variants possess mutations in the other parts of their genome that may also affect pathogenesis. Of particular interest to us are the mutations present in the accessory genes, which have been shown to contribute to pathogenesis in the host through interference with innate immune signaling, among other effects on host machinery. To examine the effects of accessory protein mutations and other nonspike mutations on SARS-CoV-2 pathogenesis, we synthesized both viruses possessing deletions in the accessory genes as well as viruses where the WA-1 spike is replaced by each variant spike gene in a SARS-CoV-2/WA-1 infectious clone. We then characterized the in vitro and in vivo replication of these viruses and compared them to both WA-1 and the full variant viruses. Our work has revealed that the accessory proteins contribute to SARS-CoV-2 pathogenesis and the nonspike mutations in variants can contribute to replication of SARS-CoV-2 and pathogenesis in the host. This work suggests that while spike mutations may enhance receptor binding and entry into cells, mutations in accessory proteins may alter clinical disease presentation.
Keywords: SARS-CoV-2; coronavirus; mouse; pathogenesis; variant.
Conflict of interest statement
The authors declare no competing interest.
Figures



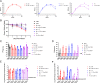

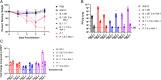

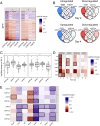
Update of
-
SARS-CoV-2 Variant Spike and accessory gene mutations alter pathogenesis.bioRxiv [Preprint]. 2022 Jun 1:2022.05.31.494211. doi: 10.1101/2022.05.31.494211. bioRxiv. 2022. Update in: Proc Natl Acad Sci U S A. 2022 Sep 13;119(37):e2204717119. doi: 10.1073/pnas.2204717119. PMID: 35677080 Free PMC article. Updated. Preprint.
References
-
- Archived: WHO timeline - COVID-19. https://www.who.int/news/item/27-04-2020-who-timeline---covid-19. Accessed 16 August 2022.
-
- WHO coronavirus (COVID-19) dashboard. https://covid19.who.int. Accessed 16 August 2022.
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

