Unfolding the mitochondrial genome structure of green semilooper (Chrysodeixis acuta Walker): An emerging pest of onion (Allium cepa L.)
- PMID: 36040876
- PMCID: PMC9426943
- DOI: 10.1371/journal.pone.0273635
Unfolding the mitochondrial genome structure of green semilooper (Chrysodeixis acuta Walker): An emerging pest of onion (Allium cepa L.)
Abstract
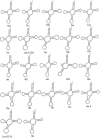
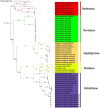
Onion is the most important crop challenged by a diverse group of insect pests in the agricultural ecosystem. The green semilooper (Chrysodeixis acuta Walker), a widespread tomato and soybean pest, has lately been described as an emergent onion crop pest in India. C. acuta whole mitochondrial genome was sequenced in this work. The circular genome of C. acuta measured 15,743 base pairs (bp) in length. Thirteen protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and one control region were found in the 37 sequence elements. With an average 395 bp gene length, the maximum and minimum gene length observed was 1749 bp and 63 bp of nad5 and trnR, respectively. Nine of the thirteen PCGs have (ATN) as a stop codon, while the other four have a single (T) as a stop codon. Except for trnS1, all of the tRNAs were capable of producing a conventional clover leaf structure. Conserved ATAGA motif sequences and poly-T stretch were identified at the start of the control region. Six overlapping areas and 18 intergenic spacer regions were found, with sizes ranged from 1 to 20 bp and 1 to 111 bp correspondingly. Phylogenetically, C. acuta belongs to the Plusiinae subfamily of the Noctuidae superfamily, and is closely linked to Trichoplusia ni species from the same subfamily. In the present study, the emerging onion pest C. acuta has its complete mitochondrial genome sequenced for the first time.
Conflict of interest statement
All the authors declare that there is no competing conflict of interest.
Figures

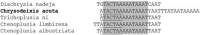

References
-
- Jadhav VG, Baviskar PP, Pathrikar DT, Bhosale GV., Export performance of Onion in India. Pharma Innov. 2022; 11:428–430.
-
- Soumia PS, Karuppaiah V, Mahajan V, Singh M. Beet Armyworm Spodoptera exigua: emerging threat to onion production. Natl. Acad. Sci. Lett. 2020; 43(5):473–6. doi: 10.1007/s40009-020-00892-5 - DOI
-
- Gedam PA, Thangasamy A, Shirsat DV, Ghosh S, Bhagat KP, Sogam OA, et al.. Screening of onion (Allium cepa L.) genotypes for drought tolerance using physiological and yield based indices through multivariate analysis. Front. Plant Sci. 2021; 12: 122. doi: 10.3389/fpls.2021.600371 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

