Reverse vaccinology approach to identify novel and immunogenic targets against Porphyromonas gingivalis: An in silico study
- PMID: 36040920
- PMCID: PMC9426909
- DOI: 10.1371/journal.pone.0273770
Reverse vaccinology approach to identify novel and immunogenic targets against Porphyromonas gingivalis: An in silico study
Abstract
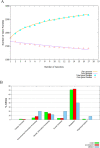
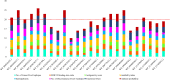
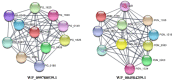
Porphyromonas gingivalis is a primary causative agent of chronic periodontitis. Moreover, it leads to several systemic diseases, including rheumatoid arthritis, cardiovascular, neurodegenerative, and Alzheimer's diseases. It seems that the development of a vaccine against this bacterium is necessary. Thus, this study decided to identify novel immunogenic targets and developed multiple epitope-based vaccines against P. gingivalis. For this purpose, the pan/core-proteome of this bacterium was studied, and the suitable vaccine targets were selected based on different properties, including exposed localization of proteins, antigenicity, non-allergenicity, non-similarity to host proteome, stability, B-cell epitopes and MHC II binding sites, sequence conservation, molecular docking, and immune simulation. Through the quartile scoring method, 12 proteins with ≥ 20 scores were considered as suitable immunogenic targets. The results of the protein domain and functional class search showed that most of the immunogenic proteins were involved in the transport and metabolism of inorganic ions and lipids. In addition, two unknown function proteins, including WP_004584259.1 and WP_099780539.1 were detected as immunogenic targets. Three constructions carrying multi-epitopes were generated including Naked, LCL, and as chimeric structures. Among them, FliC chimeric protein had the strongest affinity to the human TLR2, 4, and 6, while the LCL platform represented the highest level of immune stimulation response. The obtained results from this study revealed new insights into prophylactic routes against P. gingivalis by introducing novel immunogenic targets. However, further investigations, including site-directed mutation and immunoassay are needed to confirm the pathogenic role and protectivity of these novel proteins.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Khan S, Ali SS, Zaheer I, Saleem S, Ziaullah, Zaman N, et al. Proteome-wide mapping and reverse vaccinology-based B and T cell multi-epitope subunit vaccine designing for immune response reinforcement against Porphyromonas gingivalis. Journal of Biomolecular Structure and Dynamics. 2022;40(2):833–47. doi: 10.1080/07391102.2020.1819423 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

