Predicting COVID-19 disease severity from SARS-CoV-2 spike protein sequence by mixed effects machine learning
- PMID: 36041271
- PMCID: PMC9384346
- DOI: 10.1016/j.compbiomed.2022.105969
Predicting COVID-19 disease severity from SARS-CoV-2 spike protein sequence by mixed effects machine learning
Abstract
Epidemiological studies show that COVID-19 variants-of-concern, like Delta and Omicron, pose different risks for severe disease, but they typically lack sequence-level information for the virus. Studies which do obtain viral genome sequences are generally limited in time, location, and population scope. Retrospective meta-analyses require time-consuming data extraction from heterogeneous formats and are limited to publicly available reports. Fortuitously, a subset of GISAID, the global SARS-CoV-2 sequence repository, includes "patient status" metadata that can indicate whether a sequence record is associated with mild or severe disease. While GISAID lacks data on comorbidities relevant to severity, such as obesity and chronic disease, it does include metadata for age and sex to use as additional attributes in modeling. With these caveats, previous efforts have demonstrated that genotype-patient status models can be fit to GISAID data, particularly when country-of-origin is used as an additional feature. But are these models robust and biologically meaningful? This paper shows that, in fact, temporal and geographic biases in sequences submitted to GISAID, as well as the evolving pandemic response, particularly reduction in severe disease due to vaccination, create complex issues for model development and interpretation. This paper poses a potential solution: efficient mixed effects machine learning using GPBoost, treating country as a random effect group. Training and validation using temporally split GISAID data and emerging Omicron variants demonstrates that GPBoost models are more predictive of the impact of spike protein mutations on patient outcomes than fixed effect XGBoost, LightGBM, random forests, and elastic net logistic regression models.
Keywords: Bioinformatics; COVID-19; Machine learning; SARS-CoV-2; Viral genomics.
Copyright © 2022 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
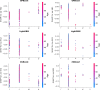
Figures







References
-
- O’Toole A., Scher E., Underwood A., Jackson B., Hill V., McCrone J.T., Colquhoun R., Ruis C., Abu-Dahab K., Taylor B., Yeats C., du Plessis L., Maloney D., Medd N., Attwood S.W., Aanensen D.M., Holmes E.C., Pybus O.G., Rambaut A. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021;7(2):veab064. - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

