Multi-omics analysis defines highly refractory RAS burdened immature subgroup of infant acute lymphoblastic leukemia
- PMID: 36042201
- PMCID: PMC9427775
- DOI: 10.1038/s41467-022-32266-4
Multi-omics analysis defines highly refractory RAS burdened immature subgroup of infant acute lymphoblastic leukemia
Abstract
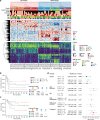
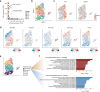
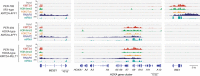
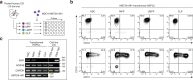
KMT2A-rearranged infant acute lymphoblastic leukemia (ALL) represents the most refractory type of childhood leukemia. To uncover the molecular heterogeneity of this disease, we perform RNA sequencing, methylation array analysis, whole exome and targeted deep sequencing on 84 infants with KMT2A-rearranged leukemia. Our multi-omics clustering followed by single-sample and single-cell inference of hematopoietic differentiation establishes five robust integrative clusters (ICs) with different master transcription factors, fusion partners and corresponding stages of B-lymphopoietic and early hemato-endothelial development: IRX-type differentiated (IC1), IRX-type undifferentiated (IC2), HOXA-type MLLT1 (IC3), HOXA-type MLLT3 (IC4), and HOXA-type AFF1 (IC5). Importantly, our deep mutational analysis reveals that the number of RAS pathway mutations predicts prognosis and that the most refractory subgroup of IC2 possesses 100% frequency and the heaviest burden of RAS pathway mutations. Our findings highlight the previously under-appreciated intra- and inter-patient heterogeneity of KMT2A-rearranged infant ALL and provide a rationale for the future development of genomics-guided risk stratification and individualized therapy.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures