Deconvoluting complex correlates of COVID-19 severity with a multi-omic pandemic tracking strategy
- PMID: 36042219
- PMCID: PMC9426371
- DOI: 10.1038/s41467-022-32397-8
Deconvoluting complex correlates of COVID-19 severity with a multi-omic pandemic tracking strategy
Abstract
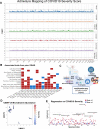
The SARS-CoV-2 pandemic has differentially impacted populations across race and ethnicity. A multi-omic approach represents a powerful tool to examine risk across multi-ancestry genomes. We leverage a pandemic tracking strategy in which we sequence viral and host genomes and transcriptomes from nasopharyngeal swabs of 1049 individuals (736 SARS-CoV-2 positive and 313 SARS-CoV-2 negative) and integrate them with digital phenotypes from electronic health records from a diverse catchment area in Northern California. Genome-wide association disaggregated by admixture mapping reveals novel COVID-19-severity-associated regions containing previously reported markers of neurologic, pulmonary and viral disease susceptibility. Phylodynamic tracking of consensus viral genomes reveals no association with disease severity or inferred ancestry. Summary data from multiomic investigation reveals metagenomic and HLA associations with severe COVID-19. The wealth of data available from residual nasopharyngeal swabs in combination with clinical data abstracted automatically at scale highlights a powerful strategy for pandemic tracking, and reveals distinct epidemiologic, genetic, and biological associations for those at the highest risk.
© 2022. The Author(s).
Conflict of interest statement
V.N.P. is a consultant and scientific advisor for Biomarin. A.G.I. is a founder of Galatea Bio. C.A.B. is on the SAB of Catamaran Bio and DeepCell, Inc. AJR is a Scientific Advisor of Merck, MTW is a stockholder of Personalis, Inc., G.P.S. is on the SAB of Jumpcode Genomics. C.D.B. is founder and CEO of Galatea Bio and a SAB member for Genomelink, Etalon Dx, and Embark Vet. M.A.R. is on the SAB of 54Gene and Related Sciences, is scientific founder of Broadwing Bio, and has advised BioMarin, Third Rock Ventures, and MazeTx. E.A.A. is a founder of Personalis, Inc, DeepCell, Inc, and Svexa Inc., a founding advisor of Nuevocor, a non-executive director at AstraZeneca, and an advisor to SequenceBio, Novartis, Medical Excellence Capital, Foresite Capital, and Third Rock Ventures. The remaining authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

