Single-cell transcriptome and translatome dual-omics reveals potential mechanisms of human oocyte maturation
- PMID: 36042231
- PMCID: PMC9427852
- DOI: 10.1038/s41467-022-32791-2
Single-cell transcriptome and translatome dual-omics reveals potential mechanisms of human oocyte maturation
Abstract
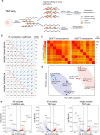
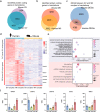
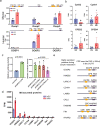
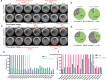
The combined use of transcriptome and translatome as indicators of gene expression profiles is usually more accurate than the use of transcriptomes alone, especially in cell types governed by translational regulation, such as mammalian oocytes. Here, we developed a dual-omics methodology that includes both transcriptome and translatome sequencing (T&T-seq) of single-cell oocyte samples, and we used it to characterize the transcriptomes and translatomes during mouse and human oocyte maturation. T&T-seq analysis revealed distinct translational expression patterns between mouse and human oocytes and delineated a sequential gene expression regulation from the cytoplasm to the nucleus during human oocyte maturation. By these means, we also identified a functional role of OOSP2 inducing factor in human oocyte maturation, as human recombinant OOSP2 induced in vitro maturation of human oocytes, which was blocked by anti-OOSP2. Single-oocyte T&T-seq analyses further elucidated that OOSP2 induces specific signaling pathways, including small GTPases, through translational regulation.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. - PubMed
-
- Broix L, Turchetto S, Nguyen L. Coordination between transport and local translation in neurons. Trends Cell Biol. 2021;31:372–386. - PubMed
-
- Piccirillo CA, Bjur E, Topisirovic I, Sonenberg N, Larsson O. Translational control of immune responses: from transcripts to translatomes. Nat. Immunol. 2014;15:503–511. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

