Fiber photometry in striatum reflects primarily nonsomatic changes in calcium
- PMID: 36042311
- PMCID: PMC10152879
- DOI: 10.1038/s41593-022-01152-z
Fiber photometry in striatum reflects primarily nonsomatic changes in calcium
Abstract
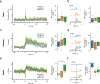
Fiber photometry enables recording of population neuronal calcium dynamics in awake mice. While the popularity of fiber photometry has grown in recent years, it remains unclear whether photometry reflects changes in action potential firing (that is, 'spiking') or other changes in neuronal calcium. In microscope-based calcium imaging, optical and analytical approaches can help differentiate somatic from neuropil calcium. However, these approaches cannot be readily applied to fiber photometry. As such, it remains unclear whether the fiber photometry signal reflects changes in somatic calcium, changes in nonsomatic calcium or a combination of the two. Here, using simultaneous in vivo extracellular electrophysiology and fiber photometry, along with in vivo endoscopic one-photon and two-photon calcium imaging, we determined that the striatal fiber photometry does not reflect spiking-related changes in calcium and instead primarily reflects nonsomatic changes in calcium.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures
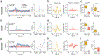
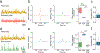

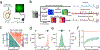


Similar articles
-
Coordinated Ramping of Dorsal Striatal Pathways preceding Food Approach and Consumption.J Neurosci. 2018 Apr 4;38(14):3547-3558. doi: 10.1523/JNEUROSCI.2693-17.2018. Epub 2018 Mar 9. J Neurosci. 2018. PMID: 29523623 Free PMC article.
-
Spectrally Resolved Fiber Photometry for Multi-component Analysis of Brain Circuits.Neuron. 2018 May 16;98(4):707-717.e4. doi: 10.1016/j.neuron.2018.04.012. Epub 2018 May 3. Neuron. 2018. PMID: 29731250 Free PMC article.
-
Simultaneous GCaMP6-based fiber photometry and fMRI in rats.J Neurosci Methods. 2017 Sep 1;289:31-38. doi: 10.1016/j.jneumeth.2017.07.002. Epub 2017 Jul 4. J Neurosci Methods. 2017. PMID: 28687521 Free PMC article.
-
Lights, fiber, action! A primer on in vivo fiber photometry.Neuron. 2024 Mar 6;112(5):718-739. doi: 10.1016/j.neuron.2023.11.016. Epub 2023 Dec 15. Neuron. 2024. PMID: 38103545 Free PMC article. Review.
-
Calcium Imaging in Vivo: How to Correctly Select and Apply Fiber Optic Photometric Indicators.Organogenesis. 2025 Dec;21(1):2489667. doi: 10.1080/15476278.2025.2489667. Epub 2025 Apr 5. Organogenesis. 2025. PMID: 40186873 Free PMC article. Review.
Cited by
-
Ventral pallidum GABA and glutamate neurons drive approach and avoidance through distinct modulation of VTA cell types.Nat Commun. 2024 May 18;15(1):4233. doi: 10.1038/s41467-024-48340-y. Nat Commun. 2024. PMID: 38762463 Free PMC article.
-
Dissociable effects of oxycodone on behavior, calcium transient activity, and excitability of dorsolateral striatal neurons.Front Neural Circuits. 2022 Oct 26;16:983323. doi: 10.3389/fncir.2022.983323. eCollection 2022. Front Neural Circuits. 2022. PMID: 36389179 Free PMC article.
-
Unraveling the Neural Circuits: Techniques, Opportunities and Challenges in Epilepsy Research.Cell Mol Neurobiol. 2024 Mar 6;44(1):27. doi: 10.1007/s10571-024-01458-5. Cell Mol Neurobiol. 2024. PMID: 38443733 Free PMC article. Review.
-
Continuous cholinergic-dopaminergic updating in the nucleus accumbens underlies approaches to reward-predicting cues.Nat Commun. 2022 Dec 24;13(1):7924. doi: 10.1038/s41467-022-35601-x. Nat Commun. 2022. PMID: 36564387 Free PMC article.
-
Fiber photometry-based investigation of brain function and dysfunction.Neurophotonics. 2024 Sep;11(Suppl 1):S11502. doi: 10.1117/1.NPh.11.S1.S11502. Epub 2023 Nov 14. Neurophotonics. 2024. PMID: 38077295 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

