The Ntan1 gene is expressed in perineural glia and neurons of adult Drosophila
- PMID: 36042338
- PMCID: PMC9427837
- DOI: 10.1038/s41598-022-18999-8
The Ntan1 gene is expressed in perineural glia and neurons of adult Drosophila
Abstract
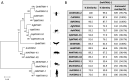
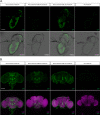
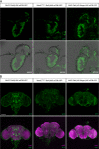
The Drosophila Ntan1 gene encodes an N-terminal asparagine amidohydrolase that we show is highly conserved throughout evolution. Protein isoforms share more than 72% of similarity with their human counterparts. At the cellular level, this gene regulates the type of glial cell growth in Drosophila larvae by its different expression levels. The Drosophila Ntan1 gene has 4 transcripts that encode 2 protein isoforms. Here we describe that although this gene is expressed at all developmental stages and adult organs tested (eye, antennae and brain) there are some transcript-dependent specificities. Therefore, both quantitative and qualitative cues could account for gene function. However, widespread developmental stage and organ-dependent expression could be masking cell-specific constraints that can be explored in Drosophila by using Gal4 drivers. We report a new genetic driver within this gene, Mz317-Gal4, that recapitulates the Ntan1 gene expression pattern in adults. It shows specific expression for perineural glia in the olfactory organs but mixed expression with some neurons in the adult brain. Memory and social behavior disturbances in mice and cancer and schizophrenia in humans have been linked to the Ntan1 gene. Therefore, these new tools in Drosophila may contribute to our understanding of the cellular basis of these alterations.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

