Adipocytes control food intake and weight regain via Vacuolar-type H+ ATPase
- PMID: 36042358
- PMCID: PMC9427743
- DOI: 10.1038/s41467-022-32764-5
Adipocytes control food intake and weight regain via Vacuolar-type H+ ATPase
Abstract
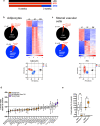
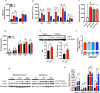
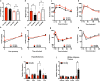
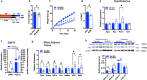
Energy metabolism becomes dysregulated in individuals with obesity and many of these changes persist after weight loss and likely play a role in weight regain. In these studies, we use a mouse model of diet-induced obesity and weight loss to study the transcriptional memory of obesity. We found that the 'metabolic memory' of obesity is predominantly localized in adipocytes. Utilizing a C. elegans-based food intake assay, we identify 'metabolic memory' genes that play a role in food intake regulation. We show that expression of ATP6v0a1, a subunit of V-ATPase, is significantly induced in both obese mouse and human adipocytes that persists after weight loss. C. elegans mutants deficient in Atp6v0A1/unc32 eat less than WT controls. Adipocyte-specific Atp6v0a1 knockout mice have reduced food intake and gain less weight in response to HFD. Pharmacological disruption of V-ATPase assembly leads to decreased food intake and less weight re-gain. In summary, using a series of genetic tools from invertebrates to vertebrates, we identify ATP6v0a1 as a regulator of peripheral metabolic memory, providing a potential target for regulation of food intake, weight loss maintenance and the treatment of obesity.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
The different functions of V-ATPase subunits in adipocyte differentiation and their expression in obese mice.Biochem Biophys Res Commun. 2024 Nov 12;733:150733. doi: 10.1016/j.bbrc.2024.150733. Epub 2024 Sep 24. Biochem Biophys Res Commun. 2024. PMID: 39332157
-
HOX-7 suppresses body weight gain and adipogenesis-related gene expression in high-fat-diet-induced obese mice.BMC Complement Altern Med. 2014 Dec 17;14:505. doi: 10.1186/1472-6882-14-505. BMC Complement Altern Med. 2014. PMID: 25515293 Free PMC article.
-
Obesity-induced changes in lipid mediators persist after weight loss.Int J Obes (Lond). 2018 Apr;42(4):728-736. doi: 10.1038/ijo.2017.266. Epub 2017 Nov 1. Int J Obes (Lond). 2018. PMID: 29089614 Free PMC article.
-
Adipocyte-specific gp130 signalling mediates exercise-induced weight reduction.Int J Obes (Lond). 2020 Mar;44(3):707-714. doi: 10.1038/s41366-019-0444-7. Epub 2019 Aug 29. Int J Obes (Lond). 2020. PMID: 31467419
-
Vacuolar (H+)-ATPases in Caenorhabditis elegans: what can we learn about giant H+ pumps from tiny worms?Biochim Biophys Acta. 2010 Oct;1797(10):1687-95. doi: 10.1016/j.bbabio.2010.07.004. Epub 2010 Jul 15. Biochim Biophys Acta. 2010. PMID: 20637717 Review.
Cited by
-
The metabolic effects of resumption of a high fat diet after weight loss are sex dependent in mice.Sci Rep. 2023 Aug 14;13(1):13227. doi: 10.1038/s41598-023-40514-w. Sci Rep. 2023. PMID: 37580448 Free PMC article.
-
Myocardin reverses insulin resistance and ameliorates cardiomyopathy by increasing IRS-1 expression in a murine model of lipodystrophy caused by adipose deficiency of vacuolar H+-ATPase V0d1 subunit.Theranostics. 2024 Mar 11;14(5):2246-2264. doi: 10.7150/thno.93192. eCollection 2024. Theranostics. 2024. PMID: 38505620 Free PMC article.
-
Transcriptional and Metabolic Changes Following Repeated Fasting and Refeeding of Adipose Stem Cells Highlight Adipose Tissue Resilience.Nutrients. 2024 Dec 13;16(24):4310. doi: 10.3390/nu16244310. Nutrients. 2024. PMID: 39770930 Free PMC article.
-
Adipose tissue fibrosis: the unwanted houseguest invited by obesity.J Endocrinol. 2023 Oct 19;259(3):e230180. doi: 10.1530/JOE-23-0180. Print 2023 Dec 1. J Endocrinol. 2023. PMID: 37855264 Free PMC article. Review.
-
Weight cycling and its effects on muscle mass, sarcopenia and sarcopenic obesity.Rev Endocr Metab Disord. 2025 Apr 15. doi: 10.1007/s11154-025-09963-8. Online ahead of print. Rev Endocr Metab Disord. 2025. PMID: 40232654 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

