Decreased NHE3 expression in colon cancer is associated with DNA damage, increased inflammation and tumor growth
- PMID: 36042372
- PMCID: PMC9427942
- DOI: 10.1038/s41598-022-19091-x
Decreased NHE3 expression in colon cancer is associated with DNA damage, increased inflammation and tumor growth
Abstract
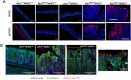
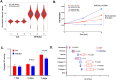
Dysregulation of intra- and extracellular pH in cancer contributes to extracellular matrix remodeling, favors cell migration, proliferation, and metastasis. Although the primary attention has been focused on the role of the ubiquitous Na+/H+ exchanger isoform NHE1, the role of NHE3, the predominant apical isoform in colonic surface epithelium in the pathogenesis of colon cancer has not been investigated. Here, we show that NHE3 mRNA expression is significantly reduced in colorectal cancer patients and that low NHE3 expression is associated with poorer survival. Deletion of NHE3 in ApcMin mice evaluated at 15 weeks of age (significant mortality was observed beyond this time) led to lower body weights, increased mucosal inflammation, increased colonic tumor numbers, evidence of enhanced DNA damage in tumor surface epithelium, and to significant alteration in the gut microbiota. In the absence of the inflammatory and microbial pressors, ca. 70% knockdown of NHE3 expression in SK-CO15 cells led to reduced intracellular pH, elevated apical pH, dramatic differences in their transcriptomic profile, increased susceptibility to DNA damage, increased proliferation, decreased apoptosis and reduced adhesion to extracellular matrix proteins. Our findings suggest that loss of NHE3 in the surface epithelium of colonic tumors has profound consequences for cancer progression and behavior.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Reshkin SJ, et al. Paclitaxel induces apoptosis via protein kinase A- and p38 mitogen-activated protein-dependent inhibition of the Na+/H+ exchanger (NHE) NHE isoform 1 in human breast cancer cells. Clin. Cancer Res. 2003;9:2366–2373. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

