Localized efficacy of environmental RNAi in Tetranychus urticae
- PMID: 36042376
- PMCID: PMC9427735
- DOI: 10.1038/s41598-022-19231-3
Localized efficacy of environmental RNAi in Tetranychus urticae
Abstract
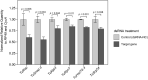
Environmental RNAi has been developed as a tool for reverse genetics studies and is an emerging pest control strategy. The ability of environmental RNAi to efficiently down-regulate the expression of endogenous gene targets assumes efficient uptake of dsRNA and its processing. In addition, its efficiency can be augmented by the systemic spread of RNAi signals. Environmental RNAi is now a well-established tool for the manipulation of gene expression in the chelicerate acari, including the two-spotted spider mite, Tetranychus urticae. Here, we focused on eight single and ubiquitously-expressed genes encoding proteins with essential cellular functions. Application of dsRNAs that specifically target these genes led to whole mite body phenotypes-dark or spotless. These phenotypes were associated with a significant reduction of target gene expression, ranging from 20 to 50%, when assessed at the whole mite level. Histological analysis of mites treated with orally-delivered dsRNAs was used to investigate the spatial range of the effectiveness of environmental RNAi. Although macroscopic changes led to two groups of body phenotypes, silencing of target genes was associated with the distinct cellular phenotypes. We show that regardless of the target gene tested, cells that displayed histological changes were those that are in direct contact with the dsRNA-containing gut lumen, suggesting that the greatest efficiency of the orally-delivered dsRNAs is localized to gut tissues in T. urticae.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Whangbo JS, Hunter CP. Environmental RNA interference. Trends Genet. 2008;24:297–305. - PubMed
-
- Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. - PubMed
-
- Fraser AG, et al. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. - PubMed
-
- Gönczy P, et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

