Crowdsourced benchmarking of taxonomic metagenome profilers: lessons learned from the sbv IMPROVER Microbiomics challenge
- PMID: 36042406
- PMCID: PMC9429340
- DOI: 10.1186/s12864-022-08803-2
Crowdsourced benchmarking of taxonomic metagenome profilers: lessons learned from the sbv IMPROVER Microbiomics challenge
Abstract
Background: Selection of optimal computational strategies for analyzing metagenomics data is a decisive step in determining the microbial composition of a sample, and this procedure is complex because of the numerous tools currently available. The aim of this research was to summarize the results of crowdsourced sbv IMPROVER Microbiomics Challenge designed to evaluate the performance of off-the-shelf metagenomics software as well as to investigate the robustness of these results by the extended post-challenge analysis. In total 21 off-the-shelf taxonomic metagenome profiling pipelines were benchmarked for their capacity to identify the microbiome composition at various taxon levels across 104 shotgun metagenomics datasets of bacterial genomes (representative of various microbiome samples) from public databases. Performance was determined by comparing predicted taxonomy profiles with the gold standard.
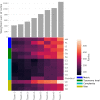
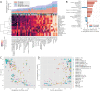
Results: Most taxonomic profilers performed homogeneously well at the phylum level but generated intermediate and heterogeneous scores at the genus and species levels, respectively. kmer-based pipelines using Kraken with and without Bracken or using CLARK-S performed best overall, but they exhibited lower precision than the two marker-gene-based methods MetaPhlAn and mOTU. Filtering out the 1% least abundance species-which were not reliably predicted-helped increase the performance of most profilers by increasing precision but at the cost of recall. However, the use of adaptive filtering thresholds determined from the sample's Shannon index increased the performance of most kmer-based profilers while mitigating the tradeoff between precision and recall.
Conclusions: kmer-based metagenomic pipelines using Kraken/Bracken or CLARK-S performed most robustly across a large variety of microbiome datasets. Removing non-reliably predicted low-abundance species by using diversity-dependent adaptive filtering thresholds further enhanced the performance of these tools. This work demonstrates the applicability of computational pipelines for accurately determining taxonomic profiles in clinical and environmental contexts and exemplifies the power of crowdsourcing for unbiased evaluation.
Keywords: Bacterial communities; Computational method benchmarking; Crowdsourcing; Metagenomics; Microbiome; Sbv IMPROVER; Taxonomic profiling.
© 2022. The Author(s).
Conflict of interest statement
All authors except V.K.N., F.M., V.K., V.C., U.M., S.K., and A.C.M. are employees of Philip Morris International. S. B and V.B. were employees of Philip Morris International at the time the work was performed. V.K.N., V.K., V.C., U.M., and S.K. are employees of Corteva Agrisciences.
Figures


References
-
- Scotti E, Boué S, Lo Sasso G, Zanetti F, Belcastro V, Poussin C, et al. Exploring the microbiome in health and disease: implications for toxicology. Toxicol Res Appl. 2017;1:2397847317741884.
MeSH terms
LinkOut - more resources
Full Text Sources

