Genome mining of Streptomyces sp. BRB081 reveals the production of the antitumor pyrrolobenzodiazepine sibiromycin
- PMID: 36043042
- PMCID: PMC9420162
- DOI: 10.1007/s13205-022-03305-0
Genome mining of Streptomyces sp. BRB081 reveals the production of the antitumor pyrrolobenzodiazepine sibiromycin
Abstract
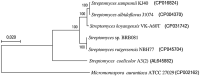
Employing a genome mining approach, this work aimed to further explore the secondary metabolism associated genes of Streptomyces sp. BRB081, a marine isolate. The genomic DNA of BRB081 was sequenced and assembled in a synteny-based pipeline for biosynthetic gene clusters (BGCs) annotation. A total of 27 BGCs were annotated, including a sibiromycin complete cluster, a bioactive compound with potent antitumor activity. The production of sibiromycin, a pyrrolobenzodiazepine, was confirmed by the analysis of obtained BRB081 extract by HPLC-MS/MS, which showed the presence of the sibiromycin ions themselves, as well as its imine and methoxylated forms. To verify the presence of this cluster in other genomes available in public databases, a genome neighborhood network (GNN) was constructed with the non-ribosomal peptide synthetase (NRPS) gene from Streptomyces sp. BRB081. Although the literature does not report the occurrence of the sibiromycin BGC in any other microorganism than Streptosporangium sibiricum, we have located this BGC in 10 other genomes besides the BRB081 isolate, all of them belonging to the Actinomycetia class. These findings strengthen the importance of uninterrupted research for new producer strains of secondary metabolites with uncommon biological activities. These results reinforced the accuracy and robustness of genomics in the screening of natural products. Furthermore, the unprecedented nature of this discovery confirms the unknown metabolic potential of the Actinobacteria phylum and the importance of continuing screening studies in this taxon.
Supplementary information: The online version contains supplementary material available at 10.1007/s13205-022-03305-0.
Keywords: Actinobacteria; Genome mining; Genomics; Secondary metabolism; Sibiromycin; Streptomyces.
© King Abdulaziz City for Science and Technology 2022.
Conflict of interest statement
Conflict of interestThe authors declare that they have no conflict of interest in the publication. This research does not involve human/ animal participants.
Figures



References
-
- Almeida EL, Carrillo RAF, Jackson SA, Dobson ADW. Comparative genomics of marine sponge-derived Streptomyces spp. isolates SM17 and SM18 with their closest terrestrial relatives provides novel insights into environmental niche adaptations and secondary metabolite biosynthesis potential. Front Microbiol. 2019 doi: 10.3389/fmicb.2019.01713. - DOI - PMC - PubMed
-
- Almeida EL, Kaur N, Jennings LK, Carrillo RAF, Jackson SA, Thomas OP, Dobson ADW. Genome mining coupled with OSMAC-based cultivation reveal differential production of Surugamide A by the marine sponge isolate Streptomyces sp. SM17 when compared to its terrestrial relative S. albidoflavus J1074. Microorganisms. 2019 doi: 10.3390/microorganisms7100394. - DOI - PMC - PubMed
-
- Benaud N, Edwards RJ, Amos TG, D'Agostino PM, Gutierrez-Chavez C, Montgomery K, Nicetic I, Ferrari BC. Antarctic desert soil bacteria exhibit high novel natural product potential, evaluated through long-read genome sequencing and comparative genomics. Environ Microbiol. 2021 doi: 10.1111/1462-2920.15300. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources

