Common Pathogens and Drug Resistance of Neonatal Pneumonia with New Multichannel Sensor
- PMID: 36043149
- PMCID: PMC9377937
- DOI: 10.1155/2022/2208636
Common Pathogens and Drug Resistance of Neonatal Pneumonia with New Multichannel Sensor
Retraction in
-
Retracted: Common Pathogens and Drug Resistance of Neonatal Pneumonia with New Multichannel Sensor.Contrast Media Mol Imaging. 2023 Jul 12;2023:9859764. doi: 10.1155/2023/9859764. eCollection 2023. Contrast Media Mol Imaging. 2023. PMID: 37476460 Free PMC article.
Abstract
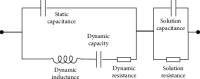
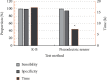
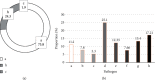
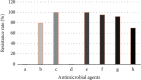
This study aimed to study the application value of a new multichannel sensor in pathogen detection and drug resistance analysis of neonatal pneumonia. 180 newborns with infectious pneumonia were selected, and a new multichannel piezoelectric sensor was constructed. The traditional Kirby-Bauer (K-B) method and the piezoelectric sensor were adopted to detect the pathogens and drug resistance in newborn samples, respectively. The results showed that the sensitivity and specificity under the K-B method (99.58% and 99.32%) and the multichannel piezoelectric sensor (99.43% and 94.29%) were not statistically different (P > 0.05). The detection time (17.25 h) of the K-B method was significantly longer than that (7.43 h) of the multichannel piezoelectric sensor (P < 0.05). From the results of pathogen detection, it was found that Klebsiella pneumoniae accounted for a relatively high proportion of 25.1%, followed by Staphylococcus aureus and Haemophilus influenzae of 13.4% and 12.33%, respectively. The resistance rate of the Staphylococcus aureus to vancomycin and rifampicin was as high as 100% and that to gentamicin, ciprofloxacin, and erythromycin reached more than 50%. In short, the new multichannel piezoelectric sensor had the high sensitivity and specificity for the pathogens' detection of neonatal pneumonia, and it required a shorter time. The pathogens were mostly Gram-negative bacteria, followed by Gram-positive bacteria and fungi. Klebsiella pneumoniae, Staphylococcus aureus, and Haemophilus influenzae were the main ones. The neonatal pneumonia pathogens had also strong drug resistance against vancomycin, rifampicin, chloramphenicol, meropenem, amikacin sulfate, chloramphenicol, and many other antibacterial drugs.
Copyright © 2022 Xueping Dong et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures



References
-
- Lee C. R., Lee J. H., Park K. S., et al. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Frontiers in Cellular and Infection Microbiology . 2017 Nov 21;7:p. 483. doi: 10.3389/fcimb.2017.00483. - DOI - PMC - PubMed
-
- Marques C., Menezes J., Belas A., et al. Klebsiella pneumoniae causing urinary tract infections in companion animals and humans: population structure, antimicrobial resistance and virulence genes. Journal of Antimicrobial Chemotherapy . 2019 Mar 1;74(3):594–602. doi: 10.1093/jac/dky499. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
