Co-amplified with PDGFRA, IGFBP7 is a prognostic biomarker correlated with the immune infiltrations of glioma
- PMID: 36043552
- PMCID: PMC9972101
- DOI: 10.1002/cam4.5187
Co-amplified with PDGFRA, IGFBP7 is a prognostic biomarker correlated with the immune infiltrations of glioma
Abstract
Background: A subgroup of glioma carry genetic 4q12 amplification including platelet derived growth factor receptor α (PDGFRA) and insulin like growth factor binding protein 7 (IGFBP7). However, the prognosis of PDGFRA and IGFBP7 in glioma is unclear.
Methods: The prognosis of PDGFRA and IGFBP7 was determined using cox regression and Kaplan-Meier survival analysis. Pathways associated with IGFBP7 were analyzed through gene set enrichment analysis (GSEA). Immune profiling of glioma was determined using "ESTIMATE" and "TIMER" database.
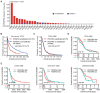
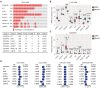
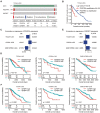
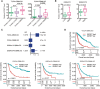
Results: PDGFRA amplification or expression was not correlated with the outcomes of glioblastoma (GBM). IGFBP7 but not PDGFRA was over-expressed in GBM. IGFBP7 over-expression was correlated with the unfavorable outcomes of GBM. In lower grade glioma (LGG), PDGFRA over-expression was not correlated with the unfavorable prognosis of LGG, while, IGFBP7 was a prognostic biomarker of LGG. LGG patients with IGFBP7 lower expressions had prolonged clinical overall survival. Combination of IDH mutation, LGG grade and IGFBP7 achieved even better prognostic effects in LGG. Moreover, IGFBP7 was over-expressed in glioma patients with wild type IDH or with high grades. IGFBP7 over-expression was correlated with the unfavorable outcomes of glioma. Furthermore, IGFBP7 was hypo-methylated in GBM or LGG patients without IDH mutations. IGFBP7 hyper-methylation was correlated with the lower overall survival of GBM or LGG. LGG patients with wild type IDH and with IGFBP7 hypo-methylation demonstrated even worse prognosis. IGFBP7 was associated with multiple immune-related signaling pathways in GBM or LGG. The stromal score, immune score and the infiltrations of immune cells were also correlated with IGFBP7 and the prognosis of LGG.
Conclusions: IGFBP7 but not PDGFRA served an ideal prognostic marker and therapeutic target of glioma.
Keywords: IGFBP7; PDGFRA; glioblastoma; immune infiltrations; lower grade glioma.
© 2022 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





Similar articles
-
RUNX1 and REXO2 are associated with the heterogeneity and prognosis of IDH wild type lower grade glioma.Sci Rep. 2021 Jun 4;11(1):11836. doi: 10.1038/s41598-021-91382-1. Sci Rep. 2021. PMID: 34088969 Free PMC article.
-
Identification of potential biomarkers related to glioma survival by gene expression profile analysis.BMC Med Genomics. 2019 Mar 20;11(Suppl 7):34. doi: 10.1186/s12920-019-0479-6. BMC Med Genomics. 2019. PMID: 30894197 Free PMC article.
-
SHOX2 is a Potent Independent Biomarker to Predict Survival of WHO Grade II-III Diffuse Gliomas.EBioMedicine. 2016 Nov;13:80-89. doi: 10.1016/j.ebiom.2016.10.040. Epub 2016 Oct 28. EBioMedicine. 2016. PMID: 27840009 Free PMC article.
-
The prognostic significance of CDKN2A homozygous deletion in IDH-mutant lower-grade glioma and glioblastoma: a systematic review of the contemporary literature.J Neurooncol. 2020 Jun;148(2):221-229. doi: 10.1007/s11060-020-03528-2. Epub 2020 May 8. J Neurooncol. 2020. PMID: 32385699
-
LAMP1/2 as potential diagnostic and prognostic marker for brain lower grade glioma: A review.Medicine (Baltimore). 2023 Aug 18;102(33):e34604. doi: 10.1097/MD.0000000000034604. Medicine (Baltimore). 2023. PMID: 37603525 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

