Identification and Characterization of the Alternative σ28 Factor in Treponema denticola
- PMID: 36043861
- PMCID: PMC9487585
- DOI: 10.1128/jb.00248-22
Identification and Characterization of the Alternative σ28 Factor in Treponema denticola
Abstract
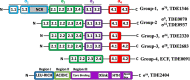
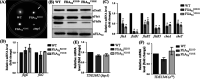
FliA (also known as σ28), a member of the bacterial σ70 family of transcription factors, directs RNA polymerase to flagellar late (class 3) promoters and initiates transcription. FliA has been studied in several bacteria, yet its role in spirochetes has not been established. In this report, we identify and functionally characterize a FliA homolog (TDE2683) in the oral spirochete Treponema denticola. Computational, genetic, and biochemical analyses demonstrated that TDE2683 has a structure similar to that of the σ28 of Escherichia coli, binds to σ28-dependent promoters, and can functionally replace the σ28 of E. coli. However, unlike its counterparts from other bacteria, TDE2683 cannot be deleted, suggesting its essential role in the survival of T. denticola. In vitro site-directed mutagenesis revealed that E221 and V231, two conserved residues in the σ4 region of σ28, are indispensable for the binding activity of TDE2683 to the σ28-dependent promoter. We then mutated these two residues in T. denticola and found that the mutations impair the expression of flagellin and chemotaxis genes and bacterial motility as well. Cryo-electron tomography analysis further revealed that the mutations disrupt the flagellar symmetry (i.e., number and placement) of T. denticola. Collectively, these results indicate that TDE2683 is a σ28 transcription factor that regulates the class 3 gene expression and controls the flagellar symmetry of T. denticola. To the best of our knowledge, this is the first report establishing the functionality of FliA in spirochetes. IMPORTANCE Spirochetes are a group of medically important but understudied bacteria. One of the unique aspects of spirochetes is that they have periplasmic flagella (PF, also known as endoflagella) which give rise to their unique spiral shape and distinct swimming behaviors and play a critical role in the pathophysiology of spirochetes. PF are structurally similar to external flagella, but the underpinning mechanism that regulates PF biosynthesis and assembly remains largely unknown. By using the oral spirochete Treponema denticola as a model, this report provides several lines of evidence that FliA, a σ28 transcriptional factor, regulates the late flagellin gene (class 3) expression, PF assembly, and flagellar symmetry as well, which provides insights into flagellar regulation and opens an avenue to investigate the role of σ28 in spirochetes.
Keywords: Treponema; flagella; motility; sigma factors; spirochetes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

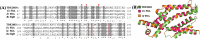



Similar articles
-
Anti-σ28 Factor FlgM Regulates Flagellin Gene Expression and Flagellar Polarity of Treponema denticola.J Bacteriol. 2023 Feb 22;205(2):e0046322. doi: 10.1128/jb.00463-22. Epub 2023 Jan 30. J Bacteriol. 2023. PMID: 36715541 Free PMC article.
-
Transcriptional and functional characterizations of multiple flagellin genes in spirochetes.Mol Microbiol. 2022 Sep;118(3):175-190. doi: 10.1111/mmi.14959. Epub 2022 Jul 18. Mol Microbiol. 2022. PMID: 35776658 Free PMC article.
-
Functional characterization of FlgM in the regulation of flagellar synthesis and motility in Yersinia pseudotuberculosis.Microbiology (Reading). 2009 Jun;155(Pt 6):1890-1900. doi: 10.1099/mic.0.026294-0. Epub 2009 Apr 21. Microbiology (Reading). 2009. PMID: 19383707
-
Transcription Regulation of Flagellins: A Structural Perspective.Biochemistry. 2025 Feb 18;64(4):770-781. doi: 10.1021/acs.biochem.4c00791. Epub 2025 Jan 28. Biochemistry. 2025. PMID: 39874281 Review.
-
Spirochete chemotaxis, motility, and the structure of the spirochetal periplasmic flagella.Res Microbiol. 1992 Jul-Aug;143(6):597-603. doi: 10.1016/0923-2508(92)90117-7. Res Microbiol. 1992. PMID: 1475520 Review.
Cited by
-
A bipartite bacterial virulence factor targets the complement system and neutrophil activation.EMBO J. 2025 Feb;44(4):1154-1184. doi: 10.1038/s44318-024-00342-8. Epub 2025 Jan 3. EMBO J. 2025. PMID: 39753953 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

