Immune Checkpoint Blockade Outcome in Small-Cell Lung Cancer and Its Relationship With Retinoblastoma Mutation Status and Function
- PMID: 36044718
- PMCID: PMC9489185
- DOI: 10.1200/PO.22.00257
Immune Checkpoint Blockade Outcome in Small-Cell Lung Cancer and Its Relationship With Retinoblastoma Mutation Status and Function
Abstract
Purpose: Immune checkpoint blockade (ICB) in conjunction with chemotherapy is approved for the treatment of extensive-stage small-cell lung cancer (SCLC). Although specific genomic abnormalities such as KEAP1 and STK11 gene mutations are associated with resistance to ICB in non-SCLC, no genomic abnormality has been found in association with resistance to ICB in SCLC.
Materials and methods: We first analyzed a retrospective cohort of 42 patients with SCLC treated with single-agent ICB or ICB combination (data set A). We then validated our results in a large prospective clinical trial of 460 patients (CheckMate 032, data set B). DNA and RNA sequencing were performed.
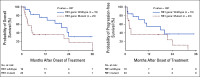
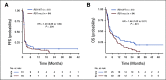
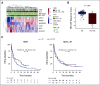
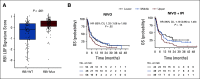
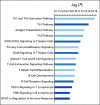
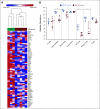
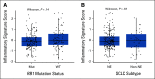
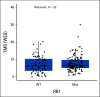
Results: In data set A, patients treated with ICB with RB1 wild-type (WT) had a median overall survival (OS) of 23.1 months (95% CI, 9 to 37.5), whereas the RB1 mutant OS was 5 months (95% CI, 2.5 to 26; P = .04). Differentially expressed gene analysis between RB1 mutant and RB1 WT samples indicated the enrichment of downregulated immune-related genes and an immune exclusion phenotype among RB1 mutant but not in the RB1 WT tumor samples. We then assessed results from 460 patients enrolled in CheckMate 032, a trial of nivolumab (NIVO) or NIVO + ipilimumab only in SCLC. In this large cohort, RB1 WT patients had significantly improved outcome with NIVO therapy compared with mutant patients (hazard ratio, 1.41; 95% CI, 1.02 to 2.01; P = .041). High RB1 loss-of-function (LOF) signature scores significantly associated with neuroendocrine subtypes (ASCL1 and NeuroD1). However, neuroendocrine subtypes did not associate with OS. Remarkably, patients with lower RB1 LOF scores had longer OS following treatment with NIVO.
Conclusion: SCLC patients with RB1 WT status or lower RB1 LOF signature scores by transcriptomics have better outcomes with ICB monotherapy.
Conflict of interest statement
Figures


References
-
- Ellis PM, Vella ET, Ung YC. Immune checkpoint inhibitors for patients with advanced non–small-cell lung cancer: A systematic review. Clin Lung Cancer. 2017;18:444–459.e1. - PubMed
-
- Rizvi H, Sanchez-Vega F, La K, et al. Molecular determinants of response to anti–programmed cell death (PD)-1 and anti–programmed death-ligand 1 (PD-L1) blockade in patients with non–small-cell lung cancer profiled with targeted next-generation sequencing. J Clin Oncol. 2018;36:633–641. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

