Innate type 2 immunity controls hair follicle commensalism by Demodex mites
- PMID: 36044899
- PMCID: PMC9561030
- DOI: 10.1016/j.immuni.2022.08.001
Innate type 2 immunity controls hair follicle commensalism by Demodex mites
Abstract
Demodex mites are commensal parasites of hair follicles (HFs). Normally asymptomatic, inflammatory outgrowth of mites can accompany malnutrition, immune dysfunction, and aging, but mechanisms restricting Demodex outgrowth are not defined. Here, we show that control of mite HF colonization in mice required group 2 innate lymphoid cells (ILC2s), interleukin-13 (IL-13), and its receptor, IL-4Ra-IL-13Ra1. HF-associated ILC2s elaborated IL-13 that attenuated HFs and epithelial proliferation at anagen onset; in their absence, Demodex colonization led to increased epithelial proliferation and replacement of gene programs for repair by aberrant inflammation, leading to the loss of barrier function and HF exhaustion. Humans with rhinophymatous acne rosacea, an inflammatory condition associated with Demodex, had increased HF inflammation with decreased type 2 cytokines, consistent with the inverse relationship seen in mice. Our studies uncover a key role for skin ILC2s and IL-13, which comprise an immune checkpoint that sustains cutaneous integrity and restricts pathologic infestation by colonizing HF mites.
Keywords: Demodex mites; IL-13; ILC2; barrier function; hair follicle stem cell; innate immunity; rhinophyma; skin homeostasis; tissue immunity; type 2 immunity.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests R.M.L. is a member of the Scientific Resource Board at Genentech and serves on the Advisory Board at Immunity.
Figures
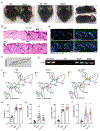

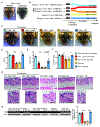
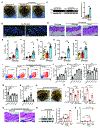


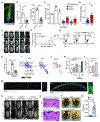
Comment in
-
A hairy situation for ILC2s.Immunity. 2022 Oct 11;55(10):1756-1758. doi: 10.1016/j.immuni.2022.09.009. Immunity. 2022. PMID: 36223723
References
-
- Anderson MJ (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecology 26, 32–46.
Publication types
MeSH terms
Substances
Grants and funding
- T35 HL007649/HL/NHLBI NIH HHS/United States
- F32 DK121476/DK/NIDDK NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- P01 HL107202/HL/NHLBI NIH HHS/United States
- R01 AI026918/AI/NIAID NIH HHS/United States
- F32 HL140868/HL/NHLBI NIH HHS/United States
- K08 AR074556/AR/NIAMS NIH HHS/United States
- H23 IP000554/IP/NCIRD CDC HHS/United States
- K08 AI159229/AI/NIAID NIH HHS/United States
- T32 AR007175/AR/NIAMS NIH HHS/United States
- R37 AI026918/AI/NIAID NIH HHS/United States
- K08 AR075880/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

