Kinetic proofreading through the multi-step activation of the ZAP70 kinase underlies early T cell ligand discrimination
- PMID: 36045187
- PMCID: PMC9477740
- DOI: 10.1038/s41590-022-01288-x
Kinetic proofreading through the multi-step activation of the ZAP70 kinase underlies early T cell ligand discrimination
Abstract
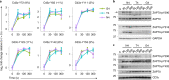
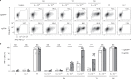
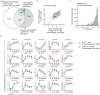
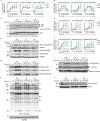
T cells recognize a few high-affinity antigens among a vast array of lower affinity antigens. According to the kinetic proofreading model, antigen discrimination properties could be explained by the gradual amplification of small differences in binding affinities as the signal is transduced downstream of the T cell receptor. Which early molecular events are affected by ligand affinity, and how, has not been fully resolved. Here, we used time-resolved high-throughput proteomic analyses to identify and quantify the phosphorylation events and protein-protein interactions encoding T cell ligand discrimination in antigen-experienced T cells. Although low-affinity ligands induced phosphorylation of the Cd3 chains of the T cell receptor and the interaction of Cd3 with the Zap70 kinase as strongly as high-affinity ligands, they failed to activate Zap70 to the same extent. As a result, formation of the signalosome of the Lat adaptor was severely impaired with low- compared with high-affinity ligands, whereas formation of the signalosome of the Cd6 receptor was affected only partially. Overall, this study provides a comprehensive map of molecular events associated with T cell ligand discrimination.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









Comment in
-
ZAP70 holds the key to kinetic proofreading for TCR ligand discrimination.Nat Immunol. 2022 Sep;23(9):1293-1294. doi: 10.1038/s41590-022-01297-w. Nat Immunol. 2022. PMID: 36045188 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

