A microbial supply chain for production of the anti-cancer drug vinblastine
- PMID: 36045295
- PMCID: PMC9452304
- DOI: 10.1038/s41586-022-05157-3
A microbial supply chain for production of the anti-cancer drug vinblastine
Abstract
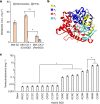
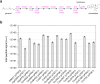
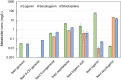
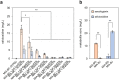
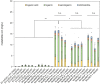
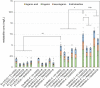
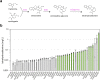
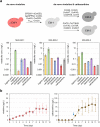
Monoterpene indole alkaloids (MIAs) are a diverse family of complex plant secondary metabolites with many medicinal properties, including the essential anti-cancer therapeutics vinblastine and vincristine1. As MIAs are difficult to chemically synthesize, the world's supply chain for vinblastine relies on low-yielding extraction and purification of the precursors vindoline and catharanthine from the plant Catharanthus roseus, which is then followed by simple in vitro chemical coupling and reduction to form vinblastine at an industrial scale2,3. Here, we demonstrate the de novo microbial biosynthesis of vindoline and catharanthine using a highly engineered yeast, and in vitro chemical coupling to vinblastine. The study showcases a very long biosynthetic pathway refactored into a microbial cell factory, including 30 enzymatic steps beyond the yeast native metabolites geranyl pyrophosphate and tryptophan to catharanthine and vindoline. In total, 56 genetic edits were performed, including expression of 34 heterologous genes from plants, as well as deletions, knock-downs and overexpression of ten yeast genes to improve precursor supplies towards de novo production of catharanthine and vindoline, from which semisynthesis to vinblastine occurs. As the vinblastine pathway is one of the longest MIA biosynthetic pathways, this study positions yeast as a scalable platform to produce more than 3,000 natural MIAs and a virtually infinite number of new-to-nature analogues.
© 2022. The Author(s).
Conflict of interest statement
J.D.K., J.Z., L.G.H., K.V., V.D., S.E.O. and M.K.J. are inventors on pending patent applications. J.D.K. has a financial interest in Amyris, Lygos, Demetrix, Napigen, Apertor Pharmaceuticals, Maple Bio, Ansa Biotechnologies, Berkeley Yeast and Zero Acre Farms. The other authors declare no competing interests.
Figures







Comment in
-
Fields to fermentors: Brewing botanical chemotherapeutic precursors using genetically engineered yeast.Med. 2022 Nov 11;3(11):727-729. doi: 10.1016/j.medj.2022.10.006. Med. 2022. PMID: 36370691
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

