Targeting TRAF3IP2 inhibits angiogenesis in glioblastoma
- PMID: 36046049
- PMCID: PMC9421153
- DOI: 10.3389/fonc.2022.893820
Targeting TRAF3IP2 inhibits angiogenesis in glioblastoma
Abstract
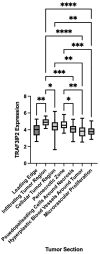
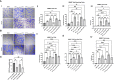
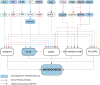
Increased vascularization, also known as neoangiogenesis, plays a major role in many cancers, including glioblastoma multiforme (GBM), by contributing to their aggressive growth and metastasis. Although anti-angiogenic therapies provide some clinical improvement, they fail to significantly improve the overall survival of GBM patients. Since various pro-angiogenic mediators drive GBM, we hypothesized that identifying targetable genes that broadly inhibit multiple pro-angiogenic mediators will significantly promote favorable outcomes. Here, we identified TRAF3IP2 (TRAF3-interacting protein 2) as a critical regulator of angiogenesis in GBM. We demonstrated that knockdown of TRAF3IP2 in an intracranial model of GBM significantly reduces vascularization. Targeting TRAF3IP2 significantly downregulated VEGF, IL6, ANGPT2, IL8, FZGF2, PGF, IL1β, EGF, PDGFRB, and VEGFR2 expression in residual tumors. Our data also indicate that exogenous addition of VEGF partially restores angiogenesis by TRAF3IP2-silenced cells, suggesting that TRAF3IP2 promotes angiogenesis through VEGF- and non-VEGF-dependent mechanisms. These results indicate the anti-angiogenic and anti-tumorigenic potential of targeting TRAF3IP2 in GBM, a deadly cancer with limited treatment options.
Keywords: Glioblastoma multiforme; TRAF3IP2; angiogenesis; inflammation; tumor microenvironment.
Copyright © 2022 Izadpanah, Daneshimehr, Willingham, Barabadi, Braun, Dumont, Mostany, Chandrasekar, Alt and Izadpanah.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


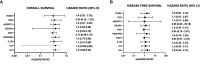
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

