Linkage QTL Mapping and Genome-Wide Association Study on Resistance in Chickpea to Pythium ultimum
- PMID: 36046237
- PMCID: PMC9420999
- DOI: 10.3389/fgene.2022.945787
Linkage QTL Mapping and Genome-Wide Association Study on Resistance in Chickpea to Pythium ultimum
Abstract
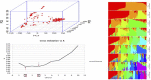
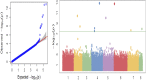
The soilborne oomycete plant pathogen Pythium ultimum causes seed rot and pre-emergence damping-off of chickpea (Cicer arietinum L.). The pathogen has been controlled for several decades using the fungicide metalaxyl as seed treatment but has re-emerged as a severe problem with the detection of metalaxyl-resistant isolates of the pathogen from infested fields in the United States Pacific Northwest. The objective of this study was to identify genetic markers and candidate genes associated with resistance to P. ultimum in an interspecific recombinant inbred line population (CRIL-7) derived from a cross between C. reticulatum (PI 599072) x C. arietinum (FLIP 84-92C) and conduct genome-wide association studies (GWAS) for disease resistance using a chickpea diversity panel consisting of 184 accessions. CRIL-7 was examined using 1029 SNP markers spanning eight linkage groups. A major QTL, "qpsd4-1," was detected on LG 4 that explained 41.8% of phenotypic variance, and a minor QTL, "qpsd8-1," was detected on LG8 that explained 4.5% of phenotypic variance. Seven candidate genes were also detected using composite interval mapping including several genes previously associated with disease resistance in other crop species. A total of 302,902 single nucleotide polymorphic (SNP) markers were used to determine population structure and kinship of the diversity panel. Marker-trait associations were established by employing different combinations of principal components (PC) and kinships (K) in the FarmCPU model. Genome-wide association studies detected 11 significant SNPs and seven candidate genes associated with disease resistance. SNP Ca4_1765418, detected by GWAS on chromosome 4, was located within QTL qpsd4-1 that was revealed in the interspecific CRIL-7 population. The present study provides tools to enable MAS for resistance to P. ultimum and identified genomic domains and candidate genes involved in the resistance of chickpea to soilborne diseases.
Keywords: Pythium; chickpea; disease; pulses; resistance.
Copyright © 2022 Agarwal, Chen, Varshney and Vandemark.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Agarwal C., Chen W., Coyne C., Vandemark G. (2020). Identifying Sources of Resistance in Chickpea to Seed Rot and Seedling Damping‐off Caused by Metalaxyl‐resistant Pythium Ultimum , [Internet]. Crop Sci. 61, 1739. [cited 2021 Jan 20];csc2.20424. 10.1002/csc2.20424 - DOI
-
- Alexander D. H., Lange K. (2011). Enhancements to the ADMIXTURE Algorithm for Individual Ancestry Estimation. BMC Bioinforma. 12 (1), 246. [cited 2020 Apr 20]Available from: https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-1... . 10.1186/1471-2105-12-246 - DOI - PMC - PubMed
-
- Bayaa B., Chen W. (2011). “Ascochyta Blight of Chickpea,” in Compendium of Chickpea and Lentil Diseases and Pests. Editors Chen W., Sharma H. C., Muehlbauer F. J. (St. Paul, MN: APS Press; ).
-
- Casas A. T., Kaiser W. J., Ingram D. M. (1990). Control of Pythium Seed Rot and Preemergence Damping-Off of Chickpea in the U.S. Pacific Northwest and Spain. Plant Dis. 74, 563–569. 10.1094/pd-74-0563 - DOI
-
- Chen W., Van Vleet S. (2016). WSU - Chickpea Damping-Off Due to Metalaxyl-Resistant Pythium: An Emerging Disease in the Palouse. [Internet]. Washington: Washingtom State University Extension. Pullman, Washington : Washington State University. Extension., 1–4. [cited 2020 Apr 20]Available from: https://pubs.wsu.edu/ItemDetail.aspx?ProductID=15853&SeriesCode=&Categor... .
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

