STArS (STrain-Amplicon-Seq), a targeted nanopore sequencing workflow for SARS-CoV-2 diagnostics and genotyping
- PMID: 36046362
- PMCID: PMC9422081
- DOI: 10.1093/biomethods/bpac020
STArS (STrain-Amplicon-Seq), a targeted nanopore sequencing workflow for SARS-CoV-2 diagnostics and genotyping
Abstract
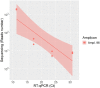
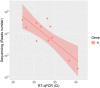
Diagnostic tests based on reverse transcription-quantitative polymerase chain reaction (RT-qPCR) are the gold standard approach to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection from clinical specimens. However, unless specifically optimized, this method is usually unable to recognize the specific viral strain responsible of coronavirus disease 2019, a crucial information that is proving increasingly important in relation to virus spread and treatment effectiveness. Even if some RT-qPCR commercial assays are currently being developed for the detection of viral strains, they focus only on single/few genetic variants that may not be sufficient to uniquely identify a specific strain. Therefore, genome sequencing approaches remain the most comprehensive solution for virus genotyping and to recognize viral strains, but their application is much less widespread due to higher costs. Starting from the well-established ARTIC protocol coupled to nanopore sequencing, in this work, we developed STArS (STrain-Amplicon-Seq), a cost/time-effective sequencing-based workflow for both SARS-CoV-2 diagnostics and genotyping. A set of 10 amplicons was initially selected from the ARTIC tiling panel, to cover: (i) all the main biologically relevant genetic variants located on the Spike gene; (ii) a minimal set of variants to uniquely identify the currently circulating strains; (iii) genomic sites usually amplified by RT-qPCR method to identify SARS-CoV-2 presence. PCR-amplified clinical samples (both positive and negative for SARS-CoV-2 presence) were pooled together with a serially diluted exogenous amplicon at known concentration and sequenced on a MinION device. Thanks to a scoring rule, STArS had the capability to accurately classify positive samples in agreement with RT-qPCR results, both at the qualitative and quantitative level. Moreover, the method allowed to effectively genotype strain-specific variants and thus also return the phylogenetic classification of SARS-CoV-2-postive samples. Thanks to the reduced turnaround time and costs, the proposed approach represents a step towards simplifying the clinical application of sequencing for viral genotyping, hopefully aiding in combatting the global pandemic.
© The Author(s) 2022. Published by Oxford University Press.
Figures

References
-
- James P, Stoddart D, Harrington E. et al. LamPORE: rapid, accurate and highly scalable molecular screening for SARS-CoV-2 infection, based on nanopore sequencing. medRxiv 2020. 10.1101/2020.08.07.20161737 (1 April 2021, date last accessed). - DOI
