Characterization of the SGLT2 Interaction Network and Its Regulation by SGLT2 Inhibitors: A Bioinformatic Analysis
- PMID: 36046822
- PMCID: PMC9421436
- DOI: 10.3389/fphar.2022.901340
Characterization of the SGLT2 Interaction Network and Its Regulation by SGLT2 Inhibitors: A Bioinformatic Analysis
Abstract
Background: Sodium-glucose cotransporter 2 (SGLT2), also known as solute carrier family 5 member 2 (SLC5A2), is a promising target for a new class of drugs primarily established as kidney-targeting, effective glucose-lowering agents used in diabetes mellitus (DM) patients. Increasing evidence indicates that besides renal effects, SGLT2 inhibitors (SGLT2i) have also a systemic impact via indirectly targeting the heart and other tissues. Our hypothesis states that the pleiotropic effects of SGLT2i are associated with their binding force, location of targets in the SGLT2 networks, targets involvement in signaling pathways, and their tissue-specific expression. Methods: Thus, to investigate differences in SGLT2i impact on human organisms, we re-created the SGLT2 interaction network incorporating its inhibitors and metformin and analyzed its tissue-specific expression using publicly available datasets. We analyzed it in the context of the so-called key terms ( autophagy, oxidative stress, aging, senescence, inflammation, AMPK pathways, and mTOR pathways) which seem to be crucial to elucidating the SGLT2 role in a variety of clinical manifestations. Results: Analysis of SGLT2 and its network components' expression confidence identified selected organs in the following order: kidney, liver, adipose tissue, blood, heart, muscle, intestine, brain, and artery according to the TISSUES database. Drug repurposing analysis of known SGLT2i pointed out the influence of SGLT1 regulators on the heart and intestine tissue. Additionally, dapagliflozin seems to also have a stronger impact on brain tissue through the regulation of SGLT3 and SLC5A11. The shortest path analysis identified interaction SIRT1-SGLT2 among the top five interactions across six from seven analyzed networks associated with the key terms. Other top first-level SGLT2 interactors associated with key terms were not only ADIPOQ, INS, GLUT4, ACE, and GLUT1 but also less recognized ILK and ADCY7. Among other interactors which appeared in multiple shortest-path analyses were GPT, COG2, and MGAM. Enrichment analysis of SGLT2 network components showed the highest overrepresentation of hypertensive disease, DM-related diseases for both levels of SGLT2 interactors. Additionally, for the extended SGLT2 network, we observed enrichment in obesity (including SGLT1), cancer-related terms, neuroactive ligand-receptor interaction, and neutrophil-mediated immunity. Conclusion: This study provides comprehensive and ranked information about the SGLT2 interaction network in the context of tissue expression and can help to predict the clinical effects of the SGLT2i.
Keywords: AMPK; SGLT2; SGLT2i; bioinformatic analysis; gene target interaction; mTOR; network; prognosis.
Copyright © 2022 Wicik, Nowak, Jarosz-Popek, Wolska, Eyileten, Siller-Matula, von Lewinski, Sourij, Filipiak and Postuła.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


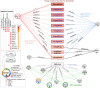


References
-
- Bae J. H., Park E. G., Kim S., Kim S. G., Hahn S., Kim N. H. (2019). Effects of Sodium-Glucose Cotransporter 2 Inhibitors on Renal Outcomes in Patients with Type 2 Diabetes: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Sci. Rep. 9, 13009. 10.1038/s41598-019-49525-y - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

