Broader Epstein-Barr virus-specific T cell receptor repertoire in patients with multiple sclerosis
- PMID: 36048016
- PMCID: PMC9437111
- DOI: 10.1084/jem.20220650
Broader Epstein-Barr virus-specific T cell receptor repertoire in patients with multiple sclerosis
Erratum in
-
Correction: Broader Epstein-Barr virus-specific T cell receptor repertoire in patients with multiple sclerosis.J Exp Med. 2022 Nov 7;219(11):e2022065010252022c. doi: 10.1084/jem.2022065010252022c. Epub 2022 Oct 28. J Exp Med. 2022. PMID: 36305889 Free PMC article. No abstract available.
Abstract
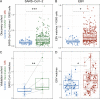
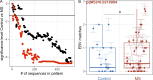
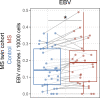
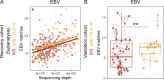
Epstein-Barr virus (EBV) infection precedes multiple sclerosis (MS) pathology and cross-reactive antibodies might link EBV infection to CNS autoimmunity. As an altered anti-EBV T cell reaction was suggested in MS, we queried peripheral blood T cell receptor β chain (TCRβ) repertoires of 1,395 MS patients, 887 controls, and 35 monozygotic, MS-discordant twin pairs for multimer-confirmed, viral antigen-specific TCRβ sequences. We detected more MHC-I-restricted EBV-specific TCRβ sequences in MS patients. Differences in genetics or upbringing could be excluded by validation in monozygotic twin pairs discordant for MS. Anti-VLA-4 treatment amplified this observation, while interferon β- or anti-CD20 treatment did not modulate EBV-specific T cell occurrence. In healthy individuals, EBV-specific CD8+ T cells were of an effector-memory phenotype in peripheral blood and cerebrospinal fluid. In MS patients, cerebrospinal fluid also contained EBV-specific central-memory CD8+ T cells, suggesting recent priming. Therefore, MS is not only preceded by EBV infection, but also associated with broader EBV-specific TCR repertoires, consistent with an ongoing anti-EBV immune reaction in MS.
© 2022 Schneider-Hohendorf et al.
Conflict of interest statement
Disclosures: R. Gittelman reported personal fees from Adaptive Biotechnologies during the conduct of the study. F. Rubelt reported a patent to US10731212B2 issued and a patent to immunoPETE related pending; and is an employee of Roche Diagnostics and receives salary, stock, and options as part of his employment compensation. C. Raposo reported being an employee and shareholder of F. Hoffmann-La Roche. B. Tackenberg reported other from F. Hoffmann-LaRoche outside the submitted work; and is a full-time employee of F. Hoffmann-LaRoche. T. Kümpfel reported personal fees from Novartis Pharma, Roche Pharma, Alexion/AstraZeneca, and Biogen for advisory boards/speaker honoraria outside the submitted work. C.C. Gross reported grants from DFG SFB/TR128 A09 during the conduct of the study; grants from DFG (single grant GR3946-3/1), IZKF (grant Kl13_010_19), Horizon2020 ReSToRe, Biogen, Roche, and Novartis Pharma; personal fees from MyLan and DIU Dresden International University GmbH; and other from Biogen, Euroimmun, MyLan, and Novartis Pharma outside the submitted work. I. Kaplan reported being an employee at Adaptive Biotechnologies during the time of this work. D. Brassat reported being a current Hoffman-La Roche employee. M. Kerschensteiner reported grants from Sanofi and personal fees from Sanofi, Biogen, Merck, Teva, Novartis, and Roche outside the submitted work. L. Klotz reported personal fees from Alexion, Bayer, Biogen, Celgene, Sanofi, Horizon, Grifols, Merck Serono, Novartis, Roche, Santhera, and Teva; and grants from German Research Foundation, IZKF Münster, IMF Münster, Biogen, Immunic AG, Novartis, and Merck Serono outside the submitted work. J.D. Lünemann reported personal fees from Abbvie, Alexion, Biogen, Novartis, Sanofi, and Takeda; and grants from Argenx, Merck, and Roche outside the submitted work. R. Liblau reported grants from GlaxoSmithKline, Foundation Bristol-Myers Squibb, Agence Nationale de la Recherche, the French MS Foundation, Cancer Research Institute, French Cancer Research Foundation, ERA-Net Narcomics, Recherche Hospitalo-Universitaire-BETPSY, and Roche; personal fees from Novartis, Sanofi-Genzyme, Biogen, Merck-Serono, and Third Rock Ventures; and other from Population Bio, Inc outside the submitted work. H. Wiendl reported personal fees for Abbvie, Alexion, Argenx, Biogen, Bristol Myers Squibb/Celgene, EMD Serono, F. Hoffmann-La Roche Ltd., Fondazione Cariplo, Genzyme, Gossamer Bio, Idorsia, Immunic, Immunovant, Janssen, Lundbeck, Merck, Neurodiem, NexGen, Novartis, PSI CRO, Roche Pharma AG, Sanofi, Swiss Multiple Sclerosis Society TEVA, UCB Biopharma, WebMD Global, and Worldwide Clinical Trials outside the submitted work. He reported grants by the DFG (CRC128 A09 and 445569437) during the conduct of the study, and funding by German Federal Ministry for Education and Research (BMBF), Deutsche Myasthenie Gesellschaft e.V., Alexion, Amicus Therapeutics Inc., Argenx, Biogen, CSL Behring, Roche, Genzyme, Merck, Novartis Pharma, Roche Pharma, and UCB Biopharma outside of the submitted work. N. Schwab reported grants from DFG, Biogen, and Roche during the conduct of the study. No other disclosures were reported.
Figures



References
-
- Abrahamyan, S., Eberspacher B., Hoshi M.M., Aly L., Luessi F., Groppa S., Klotz L., Meuth S.G., Schroeder C., Gruter T., et al. 2020. Complete Epstein-Barr virus seropositivity in a large cohort of patients with early multiple sclerosis. J. Neurol. Neurosurg. Psychiatr. 91:681–686. 10.1136/jnnp-2020-322941 - DOI - PMC - PubMed
-
- Angelini, D.F., Serafini B., Piras E., Severa M., Coccia E.M., Rosicarelli B., Ruggieri S., Gasperini C., Buttari F., Centonze D., et al. 2013. Increased CD8+ T cell response to epstein-barr virus lytic antigens in the active phase of multiple sclerosis. PLoS Pathog. 9:e1003220. 10.1371/journal.ppat.1003220 - DOI - PMC - PubMed
-
- Bagaev, D.V., Vroomans R.M.A., Samir J., Stervbo U., Rius C., Dolton G., Greenshields-Watson A., Attaf M., Egorov E.S., Zvyagin I.V., et al. 2020. VDJdb in 2019: Database extension, new analysis infrastructure and a T-cell receptor motif compendium. Nucleic Acids Res. 48:D1057–D1062. 10.1093/nar/gkz874 - DOI - PMC - PubMed
-
- Bar-Or, A., Pender M.P., Khanna R., Steinman L., Hartung H.P., Maniar T., Croze E., Aftab B.T., Giovannoni G., and Joshi M.A.. 2021. Epstein-barr virus in multiple sclerosis: Theory and emerging immunotherapies: (Trends in molecular medicine, 26:3 p:296-310, 2020). Trends Mol. Med. 27:410–411. 10.1016/j.molmed.2021.01.004 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

