CRISPRthripsis: The Risk of CRISPR/Cas9-induced Chromothripsis in Gene Therapy
- PMID: 36048170
- PMCID: PMC9585945
- DOI: 10.1093/stcltm/szac064
CRISPRthripsis: The Risk of CRISPR/Cas9-induced Chromothripsis in Gene Therapy
Abstract
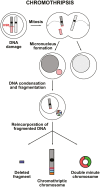
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9 nuclease system has allowed the generation of disease models and the development of therapeutic approaches for many genetic and non-genetic disorders. However, the generation of large genomic rearrangements has raised safety concerns for the clinical application of CRISPR/Cas9 nuclease approaches. Among these events, the formation of micronuclei and chromosome bridges due to chromosomal truncations can lead to massive genomic rearrangements localized to one or few chromosomes. This phenomenon, known as chromothripsis, was originally described in cancer cells, where it is believed to be caused by defective chromosome segregation during mitosis or DNA double-strand breaks. Here, we will discuss the factors influencing CRISPR/Cas9-induced chromothripsis, hereafter termed CRISPRthripsis, and its outcomes, the tools to characterize these events and strategies to minimize them.
Keywords: CRISPR/Cas9; chromosomal instability; chromothripsis; gene therapy; genome editing; genotoxicity; micronuclei.
© The Author(s) 2022. Published by Oxford University Press.
Figures