Severe Mycobacterial Immune Reconstitution Inflammatory Syndrome (IRIS) in Advanced Human Immunodeficiency Virus (HIV) Has Features of Hemophagocytic Lymphohistiocytosis and Requires Prolonged Immune Suppression
- PMID: 36048425
- PMCID: PMC10169423
- DOI: 10.1093/cid/ciac717
Severe Mycobacterial Immune Reconstitution Inflammatory Syndrome (IRIS) in Advanced Human Immunodeficiency Virus (HIV) Has Features of Hemophagocytic Lymphohistiocytosis and Requires Prolonged Immune Suppression
Abstract
Background: People with HIV and mycobacterial infections can develop immune reconstitution inflammatory syndrome (IRIS) after starting antiretroviral therapy (ART). Severe mycobacterial IRIS has an overlapping clinical phenotype with hemophagocytic lymphohistiocytosis (HLH). We evaluated the pathophysiologic similarities between mycobacterial IRIS and HLH to identify clinical and immune predictors of mycobacterial IRIS severity.
Methods: HLH criteria were applied to a longitudinal cohort of 80 patients with HIV (CD4 <100 cells/µL) and mycobacterial infections. Participants were subdivided into IRIS meeting HLH criteria (HLH-IRIS), IRIS without HLH (IRIS), and those without IRIS (non-IRIS). Clinical outcomes were evaluated by regression analyses. Soluble biomarkers and T-cell subsets were assessed at baseline and IRIS-equivalent time points.
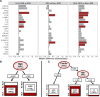
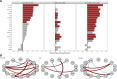
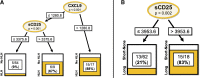
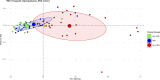
Results: HLH-IRIS patients required corticosteroids more frequently (OR: 21.5; 95%CI: 5.6-114.8) and for longer duration (21.2; 95%CI: 10.7-31.7 weeks) than those not meeting HLH criteria. Utilizing decision tree analyses, hemoglobin <9.2 g/dL was the best predictor of HLH-IRIS before ART, whereas ferritin, CXCL9 and sCD25 were most diagnostic for HLH at IRIS onset. At the IRIS timepoint, but not baseline, HLH-IRIS patients had lower regulatory and higher activated T cells along with greater production of IFNγ-IL-18 axis biomarkers compared with both IRIS and non-IRIS groups. Principal component analysis corroborated the distinct clustering of HLH-IRIS patients.
Conclusions: Severe mycobacterial IRIS and HLH have an overlapping pathogenesis involving IFNγ and unopposed T-cell activation causing severe inflammatory disease clinically distinguished by hyperferritinemia (hyperferritinemic IRIS [FIRIS]). Hemoglobin, ferritin, CXCL9, and sCD25 identify high-risk patients and may improve risk stratification and therapeutic strategies for mycobacterial IRIS.
Keywords: human immunodeficiency virus; immune reconstitution inflammatory syndrome; macrophage activation syndrome; mycobacteria; tuberculosis.
Published by Oxford University Press on behalf of Infectious Diseases Society of America 2022.
Conflict of interest statement
Potential conflicts of interest. I. S. reports a patent, unrelated to this work, “Methods for the treatment of Kaposi's sarcoma or KSHV-induced lymphoma using immunomodulatory compounds, and uses of biomarkers (WO 2016210262 A1)”. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Figures

References
-
- Benda R, David M, Franchel F, Kahn M. Multiple brain cold abscesses during pulmonary tuberculosis treated by antibiotics; proof of their tuberculous origin furnished by the surgical specimens. Bull Mem Soc Med Hop Paris 1954; 70:802–6. - PubMed
-
- World Health Organization . Consolidated guidelines on HIV prevention, testing, treatment, service delivery and monitoring: recommendations for a public health approach, 2021 update. Geneva, Switzerland: World Health Organization, 2021. - PubMed
-
- Tan HY, Yong YK, Shankar EM, et al. Aberrant inflammasome activation characterizes tuberculosis-associated immune reconstitution inflammatory syndrome. J Immunol 2016; 196:4052–63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

